nfi-I affects behavior and life-span in C. elegans but is not essential for DNA replication or survival
- PMID: 16242019
- PMCID: PMC1277823
- DOI: 10.1186/1471-213X-5-24
nfi-I affects behavior and life-span in C. elegans but is not essential for DNA replication or survival
Abstract
Background: The Nuclear Factor I (one) (NFI) family of transcription/replication factors plays essential roles in mammalian gene expression and development and in adenovirus DNA replication. Because of its role in viral DNA replication NFI has long been suspected to function in host DNA synthesis. Determining the requirement for NFI proteins in mammalian DNA replication is complicated by the presence of 4 NFI genes in mice and humans. Loss of individual NFI genes in mice cause defects in brain, lung and tooth development, but the presence of 4 homologous NFI genes raises the issue of redundant roles for NFI genes in DNA replication. No NFI genes are present in bacteria, fungi or plants. However single NFI genes are present in several simple animals including Drosophila and C. elegans, making it possible to test for a requirement for NFI in multicellular eukaryotic DNA replication and development. Here we assess the functions of the single nfi-1 gene in C. elegans.
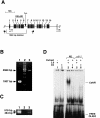
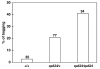
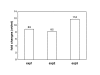
Results: C. elegans NFI protein (CeNFI) binds specifically to the same NFI-binding site recognized by vertebrate NFIs. nfi-1 encodes alternatively-spliced, maternally-inherited transcripts that are expressed at the single cell stage, during embryogenesis, and in adult muscles, neurons and gut cells. Worms lacking nfi-1 survive but have defects in movement, pharyngeal pumping and egg-laying and have a reduced life-span. Expression of the muscle gene Ce titin is decreased in nfi-1 mutant worms.
Conclusion: NFI gene function is not needed for survival in C. elegans and thus NFI is likely not essential for DNA replication in multi-cellular eukaryotes. The multiple defects in motility, egg-laying, pharyngeal pumping, and reduced lifespan indicate that NFI is important for these processes. Reduction in Ce titin expression could affect muscle function in multiple tissues. The phenotype of nfi-1 null worms indicates that NFI functions in multiple developmental and behavioral systems in C. elegans, likely regulating genes that function in motility, egg-laying, pharyngeal pumping and lifespan maintenance.
Figures






Similar articles
-
DNA-binding specificity and in vivo targets of Caenorhabditis elegans nuclear factor I.Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):12049-54. doi: 10.1073/pnas.0812894106. Epub 2009 Jul 7. Proc Natl Acad Sci U S A. 2009. PMID: 19584245 Free PMC article.
-
Lifespan extension and increased pumping rate accompany pharyngeal muscle-specific expression of nfi-1 in C. elegans.Dev Dyn. 2008 Aug;237(8):2100-7. doi: 10.1002/dvdy.21632. Dev Dyn. 2008. PMID: 18651662
-
Exon structure of the nuclear factor I DNA-binding domain from C. elegans to mammals.Mamm Genome. 1999 Apr;10(4):390-6. doi: 10.1007/s003359901008. Mamm Genome. 1999. PMID: 10087299
-
Roles of the NFI/CTF gene family in transcription and development.Gene. 2000 May 16;249(1-2):31-45. doi: 10.1016/s0378-1119(00)00140-2. Gene. 2000. PMID: 10831836 Review.
-
The Nuclear Factor I (NFI) gene family in mammary gland development and function.J Mammary Gland Biol Neoplasia. 2003 Apr;8(2):241-54. doi: 10.1023/a:1025909109843. J Mammary Gland Biol Neoplasia. 2003. PMID: 14635798 Review.
Cited by
-
Epilepsy and overgrowth-intellectual disability syndromes: a patient organization perspective on collaborating to accelerate pathways to treatment.Ther Adv Rare Dis. 2024 May 31;5:26330040241254123. doi: 10.1177/26330040241254123. eCollection 2024 Jan-Dec. Ther Adv Rare Dis. 2024. PMID: 38827639 Free PMC article. Review.
-
Evolution of glial cells: a non-bilaterian perspective.Neural Dev. 2024 Jun 21;19(1):10. doi: 10.1186/s13064-024-00184-4. Neural Dev. 2024. PMID: 38907299 Free PMC article. Review.
-
Conserved Motifs and Prediction of Regulatory Modules in Caenorhabditis elegans.G3 (Bethesda). 2012 Apr;2(4):469-81. doi: 10.1534/g3.111.001081. Epub 2012 Apr 1. G3 (Bethesda). 2012. PMID: 22540038 Free PMC article.
-
Nuclear factor I X deficiency causes brain malformation and severe skeletal defects.Mol Cell Biol. 2007 May;27(10):3855-3867. doi: 10.1128/MCB.02293-06. Mol Cell Biol. 2007. PMID: 17353270 Free PMC article.
-
DNA-binding specificity and in vivo targets of Caenorhabditis elegans nuclear factor I.Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):12049-54. doi: 10.1073/pnas.0812894106. Epub 2009 Jul 7. Proc Natl Acad Sci U S A. 2009. PMID: 19584245 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

