Specific function of a plastid sigma factor for ndhF gene transcription
- PMID: 16243785
- PMCID: PMC1266065
- DOI: 10.1093/nar/gki908
Specific function of a plastid sigma factor for ndhF gene transcription
Abstract
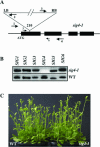
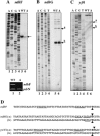
The complexity of the plastid transcriptional apparatus (two or three different RNA polymerases and numerous regulatory proteins) makes it very difficult to attribute specific function(s) to its individual components. We have characterized an Arabidopsis T-DNA insertion line disrupting the nuclear gene coding for one of the six plastid sigma factors (SIG4) that regulate the activity of the plastid-encoded RNA polymerase PEP. This mutant shows a specific diminution of transcription of the plastid ndhF gene, coding for a subunit of the plastid NDH [NAD(P)H dehydrogenase] complex. The absence of another NDH subunit, i.e. NDHH, and the absence of a chlorophyll fluorescence transient previously attributed to the activity of the plastid NDH complex indicate a strong down-regulation of NDH activity in the mutant plants. Results suggest that plastid NDH activity is regulated on the transcriptional level by an ndhF-specific plastid sigma factor, SIG4.
Figures

References
-
- Hess W.R., Börner T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999;190:1–59. - PubMed
-
- Liere K., Malliga P. Plastid RNA polymerases in higher plants. In: Aro E.-M., Andersson B., editors. Regulation of Photosynthesis. Netherlands: Kluwer Academic Publishers; 2001. pp. 29–49.
-
- Hedtke B., Börner T., Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

