Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster
- PMID: 16244705
- PMCID: PMC1262188
- DOI: 10.1371/journal.pgen.0010044
Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster
Abstract
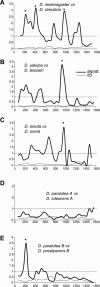
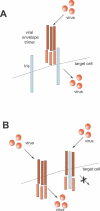
Eukaryotic genomes can usurp enzymatic functions encoded by mobile elements for their own use. A particularly interesting kind of acquisition involves the domestication of retroviral envelope genes, which confer infectious membrane-fusion ability to retroviruses. So far, these examples have been limited to vertebrate genomes, including primates where the domesticated envelope is under purifying selection to assist placental function. Here, we show that in Drosophila genomes, a previously unannotated gene (CG4715, renamed Iris) was domesticated from a novel, active Kanga lineage of insect retroviruses at least 25 million years ago, and has since been maintained as a host gene that is expressed in all adult tissues. Iris and the envelope genes from Kanga retroviruses are homologous to those found in insect baculoviruses and gypsy and roo insect retroviruses. Two separate envelope domestications from the Kanga and roo retroviruses have taken place, in fruit fly and mosquito genomes, respectively. Whereas retroviral envelopes are proteolytically cleaved into the ligand-interaction and membrane-fusion domains, Iris appears to lack this cleavage site. In the takahashii/suzukii species groups of Drosophila, we find that Iris has tandemly duplicated to give rise to two genes (Iris-A and Iris-B). Iris-B has significantly diverged from the Iris-A lineage, primarily because of the "invention" of an intron de novo in what was previously exonic sequence. Unlike domesticated retroviral envelope genes in mammals, we find that Iris has been subject to strong positive selection between Drosophila species. The rapid, adaptive evolution of Iris is sufficient to unambiguously distinguish the phylogenies of three closely related sibling species of Drosophila (D. simulans, D. sechellia, and D. mauritiana), a discriminative power previously described only for a putative "speciation gene." Iris represents the first instance of a retroviral envelope-derived host gene outside vertebrates. It is also the first example of a retroviral envelope gene that has been found to be subject to positive selection following its domestication. The unusual selective pressures acting on Iris suggest that it is an active participant in an ongoing genetic conflict. We propose a model in which Iris has "switched sides," having been recruited by host genomes to combat baculoviruses and retroviruses, which employ homologous envelope genes to mediate infection.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures








References
-
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. - PubMed
-
- Eickbush TH. Telomerase and retrotransposons: Which came first? Science. 1997;277:911–912. - PubMed
-
- Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. - PubMed
-
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. - PubMed
-
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

