Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation
- PMID: 16246912
- PMCID: PMC1266071
- DOI: 10.1093/nar/gki914
Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation
Abstract
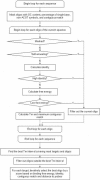
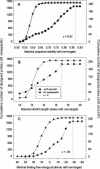
The oligonucleotide specificity for microarray hybridization can be predicted by its sequence identity to non-targets, continuous stretch to non-targets, and/or binding free energy to non-targets. Most currently available programs only use one or two of these criteria, which may choose 'false' specific oligonucleotides or miss 'true' optimal probes in a considerable proportion. We have developed a software tool, called CommOligo using new algorithms and all three criteria for selection of optimal oligonucleotide probes. A series of filters, including sequence identity, free energy, continuous stretch, GC content, self-annealing, distance to the 3'-untranslated region (3'-UTR) and melting temperature (T(m)), are used to check each possible oligonucleotide. A sequence identity is calculated based on gapped global alignments. A traversal algorithm is used to generate alignments for free energy calculation. The optimal T(m) interval is determined based on probe candidates that have passed all other filters. Final probes are picked using a combination of user-configurable piece-wise linear functions and an iterative process. The thresholds for identity, stretch and free energy filters are automatically determined from experimental data by an accessory software tool, CommOligo_PE (CommOligo Parameter Estimator). The program was used to design probes for both whole-genome and highly homologous sequence data. CommOligo and CommOligo_PE are freely available to academic users upon request.
Figures
Similar articles
-
Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy.BMC Bioinformatics. 2007 Sep 19;8:350. doi: 10.1186/1471-2105-8-350. BMC Bioinformatics. 2007. PMID: 17880708 Free PMC article.
-
A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus.BMC Genomics. 2005 Jun 17;6:95. doi: 10.1186/1471-2164-6-95. BMC Genomics. 2005. PMID: 15963225 Free PMC article.
-
OligoWiz 2.0--integrating sequence feature annotation into the design of microarray probes.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W611-5. doi: 10.1093/nar/gki399. Nucleic Acids Res. 2005. PMID: 15980547 Free PMC article.
-
[Research progress of probe design software of oligonucleotide microarrays].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2014 Feb;31(1):214-21. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2014. PMID: 24804514 Review. Chinese.
-
[Novel computerized method for designing nucleotide sequence used for DNA probes and PCR primers].Nihon Rinsho. 1994 Feb;52(2):530-41. Nihon Rinsho. 1994. PMID: 8126913 Review. Japanese.
Cited by
-
BOND: Basic OligoNucleotide Design.BMC Bioinformatics. 2013 Feb 27;14:69. doi: 10.1186/1471-2105-14-69. BMC Bioinformatics. 2013. PMID: 23444904 Free PMC article.
-
Custom design and analysis of high-density oligonucleotide bacterial tiling microarrays.PLoS One. 2009 Jun 17;4(6):e5943. doi: 10.1371/journal.pone.0005943. PLoS One. 2009. PMID: 19536279 Free PMC article.
-
An in vitro selection scheme for oligonucleotide probes to discriminate between closely related DNA sequences.Nucleic Acids Res. 2007;35(9):e66. doi: 10.1093/nar/gkm156. Epub 2007 Apr 10. Nucleic Acids Res. 2007. PMID: 17426126 Free PMC article.
-
Mismatch oligonucleotides in human and yeast: guidelines for probe design on tiling microarrays.BMC Genomics. 2008 Dec 31;9:635. doi: 10.1186/1471-2164-9-635. BMC Genomics. 2008. PMID: 19117516 Free PMC article.
-
In silico microarray probe design for diagnosis of multiple pathogens.BMC Genomics. 2008 Oct 21;9:496. doi: 10.1186/1471-2164-9-496. BMC Genomics. 2008. PMID: 18940003 Free PMC article.
References
-
- DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. - PubMed
-
- Wodicka L., Dong H., Mittmann M., Ho M.H., Lockhart D.J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 1997;15:1359–1367. - PubMed
-
- Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

