Phenotype of ENAM mutations is dosage-dependent
- PMID: 16246937
- PMCID: PMC2708095
- DOI: 10.1177/154405910508401113
Phenotype of ENAM mutations is dosage-dependent
Abstract
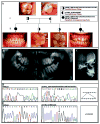
Five mutations in the ENAM gene have been found to cause hypoplastic amelogenesis imperfecta (AI), with phenotypes ranging from localized enamel pitting in carriers to severe hypoplastic AI. To determine the generality of ENAM mutations in hypoplastic AI, we sequenced the ENAM gene in ten Turkish families segregating autosomal hypoplastic AI. In two families, ENAM mutations were found. A novel nonsense mutation (g.12663C>A; p.S246X) was identified in one family segregating local hypoplastic AI as a dominant trait. Affected individuals in a second family segregating autosomal-recessive AI were compound heterozygotes for a novel insertion mutation (g.12946_12947insAGTCAGTACCAGTACTGTGTC) and a previously described insertion (g.13185_13186insAG) mutation. Heterozygous carriers of either insertion had a localized enamel-pitting phenotype. These findings substantiate that enamel phenotypes of ENAM mutations may be dose-dependent, with generalized hypoplastic AI segregating as a recessive trait and localized enamel pitting segregating as a dominant trait.
Figures


References
-
- Aldred MJ, Savarirayan R, Crawford PJM. Amelogenesis imperfecta: a classification and catalogue for the 21st century. Oral Dis. 2003;9:19–23. - PubMed
-
- Backman B. Inherited enamel defects. Ciba Found Symp. 1997;205:175–182. - PubMed
-
- Bosch C, Athanasiou AE. Landmarks, variables and norms of various numerical cephalometric analyses—cephalometric morphological and growth data references. In: Athanasiou AE, editor. Orthodontic cephalometry. London: Mosby-Wolfe; 1995. pp. 241–286.
-
- Cartwright AR, Kula K, Wright JT. Craniofacial features associated with amelogenesis imperfecta. J Craniofac Genet Dev Biol. 1999;19:148–156. - PubMed
-
- Fukae M, Tanabe T, Murakami C, Dohi N, Uchida T, Shimizu M. Primary structure of porcine 89-kDa enamelin. Adv Dent Res. 1996;10:111–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

