A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans
- PMID: 16247027
- PMCID: PMC2171198
- DOI: 10.1083/jcb.200506124
A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans
Abstract
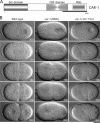
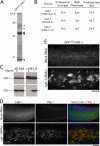
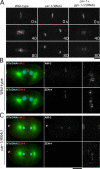
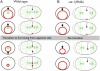
Cytokinesis completes cell division and partitions the contents of one cell to the two daughter cells. Here we characterize CAR-1, a predicted RNA binding protein that is implicated in cytokinesis. CAR-1 localizes to germline-specific RNA-containing particles and copurifies with the essential RNA helicase, CGH-1, in an RNA-dependent fashion. The atypical Sm domain of CAR-1, which directly binds RNA, is dispensable for CAR-1 localization, but is critical for its function. Inhibition of CAR-1 by RNA-mediated depletion or mutation results in a specific defect in embryonic cytokinesis. This cytokinesis failure likely results from an anaphase spindle defect in which interzonal microtubule bundles that recruit Aurora B kinase and the kinesin, ZEN-4, fail to form between the separating chromosomes. Depletion of CGH-1 results in sterility, but partially depleted worms produce embryos that exhibit the CAR-1-depletion phenotype. Cumulatively, our results suggest that CAR-1 functions with CGH-1 to regulate a specific set of maternally loaded RNAs that is required for anaphase spindle structure and cytokinesis.
Figures





References
-
- Albertson, D. 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101:61–72. - PubMed
-
- Blower, M.D., M. Nachury, R. Heald, and K. Weis. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 121:223–234. - PubMed
-
- Bringmann, H., and A.A. Hyman. 2005. A cytokinesis furrow is positioned by two consecutive signals. Nature. 436:731–734. - PubMed
-
- Cheeks, R.J., J.C. Canman, W.N. Gabriel, N. Meyer, S. Strome, and B. Goldstein. 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14:851–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

