Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning
- PMID: 16248679
- PMCID: PMC1275524
- DOI: 10.1371/journal.pbio.0030384
Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning
Abstract
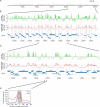
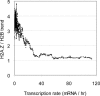
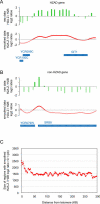
H2A.Z is an evolutionary conserved histone variant involved in transcriptional regulation, antisilencing, silencing, and genome stability. The mechanism(s) by which H2A.Z regulates these various biological functions remains poorly defined, in part due to the lack of knowledge regarding its physical location along chromosomes and the bearing it has in regulating chromatin structure. Here we mapped H2A.Z across the yeast genome at an approximately 300-bp resolution, using chromatin immunoprecipitation combined with tiling microarrays. We have identified 4,862 small regions--typically one or two nucleosomes wide--decorated with H2A.Z. Those "Z loci" are predominantly found within specific nucleosomes in the promoter of inactive genes all across the genome. Furthermore, we have shown that H2A.Z can regulate nucleosome positioning at the GAL1 promoter. Within HZAD domains, the regions where H2A.Z shows an antisilencing function, H2A.Z is localized in a wider pattern, suggesting that the variant histone regulates a silencing and transcriptional activation via different mechanisms. Our data suggest that the incorporation of H2A.Z into specific promoter-bound nucleosomes configures chromatin structure to poise genes for transcriptional activation. The relevance of these findings to higher eukaryotes is discussed.
Figures


References
-
- Khorasanizadeh S. The nucleosome: From genomic organization to genomic regulation. Cell. 2004;116:259–272. - PubMed
-
- Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003:RE4. - PubMed
-
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. - PubMed
-
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: Farewell to the tuna-can octamer? Curr Opin Genet Dev. 2004;14:165–173. - PubMed
-
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

