Evolutionarily conserved structural motifs in bacterial GST (glutathione S-transferase) are involved in protein folding and stability
- PMID: 16248855
- PMCID: PMC1385997
- DOI: 10.1042/BJ20051367
Evolutionarily conserved structural motifs in bacterial GST (glutathione S-transferase) are involved in protein folding and stability
Abstract
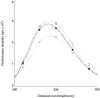
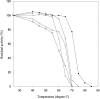
The bacterium Proteus mirabilis expresses a cytosolic class beta glutathione S-transferase (PmGST B1-1) that is part of a family of multifunctional detoxication enzymes. Like other cytosolic GSTs, PmGST B1-1 possesses two local structural motifs, an N-capping box and a hydrophobic staple motif, both of which are located between amino acids 151 and 156. The N-capping box consists of a reciprocal hydrogen bonding interaction of Thr152 with Asp155, whereas the hydrophobic staple motif consists of a hydrophobic interaction between Phe151 and Ala156. By contrast with other GSTs, PmGST B1-1 displays distinct hydrogen bond interactions in the N-capping box. In mammalian GSTs these structural elements are critical for protein folding and stability. To investigate the role played by these two motifs in a distantly related organism on the evolutionary scale, site-directed mutagenesis was used to generate several mutants of both motifs in PmGST B1-1. All mutants were efficiently overexpressed and purified, but they were quite unstable, although at different levels, indicating that protein folding was significantly destabilized. The analysis of the T152A and D155G variants indicated that the N-capping box motif plays an important role in the stability and correct folding of the enzyme. The analysis of F151A and A156G mutants revealed that the hydrophobic staple motif influences the structural maintenance of the protein and is implicated in the folding process of PmGST B1-1. Finally, the replacement of Thr152 and Asp155, as well as Phe151 and Ala156 residues influences the catalytic efficiency of the enzyme.
Figures

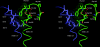
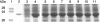
Similar articles
-
The structural roles of a conserved small hydrophobic core in the active site and an ionic bridge in domain I of Delta class glutathione S-transferase.Biochem J. 2006 Jan 1;393(Pt 1):89-95. doi: 10.1042/BJ20050555. Biochem J. 2006. PMID: 16153184 Free PMC article.
-
Contribution of the two conserved tryptophan residues to the catalytic and structural properties of Proteus mirabilis glutathione S-transferase B1-1.Biochem J. 2005 Jan 1;385(Pt 1):37-43. doi: 10.1042/BJ20040890. Biochem J. 2005. PMID: 15320869 Free PMC article.
-
A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.Structure. 1998 Jun 15;6(6):721-34. doi: 10.1016/s0969-2126(98)00074-4. Structure. 1998. PMID: 9655824
-
Enzymology of cytosolic glutathione S-transferases.Adv Pharmacol. 1994;27:37-69. doi: 10.1016/s1054-3589(08)61029-7. Adv Pharmacol. 1994. PMID: 8068560 Review. No abstract available.
-
Glutathione S-transferases, structure, regulation, and therapeutic implications.J Biol Chem. 1993 Jun 5;268(16):11475-8. J Biol Chem. 1993. PMID: 8505281 Review. No abstract available.
Cited by
-
Structural Characterization of the Xi Class Glutathione Transferase From the Haloalkaliphilic Archaeon Natrialba magadii.Front Microbiol. 2019 Jan 18;10:9. doi: 10.3389/fmicb.2019.00009. eCollection 2019. Front Microbiol. 2019. PMID: 30713525 Free PMC article.
-
A glutathione transferase from Agrobacterium tumefaciens reveals a novel class of bacterial GST superfamily.PLoS One. 2012;7(4):e34263. doi: 10.1371/journal.pone.0034263. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496785 Free PMC article.
-
Comprehensive analysis of the catalytic and structural properties of a mu-class glutathione s-transferase from Fasciola gigantica.Sci Rep. 2017 Dec 13;7(1):17547. doi: 10.1038/s41598-017-17678-3. Sci Rep. 2017. PMID: 29235505 Free PMC article.
-
The roles of highly conserved, non-catalytic residues in class A β-lactamases.Protein Sci. 2022 Jun;31(6):e4328. doi: 10.1002/pro.4328. Protein Sci. 2022. PMID: 35634774 Free PMC article.
-
Overlapping protective roles for glutathione transferase gene family members in chemical and oxidative stress response in Agrobacterium tumefaciens.Funct Integr Genomics. 2012 Mar;12(1):157-72. doi: 10.1007/s10142-011-0248-x. Epub 2011 Sep 10. Funct Integr Genomics. 2012. PMID: 21909786
References
-
- Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. - PubMed
-
- Hayes J. D., McLellan L. I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Rad. Res. 1999;31:273–300. - PubMed
-
- Armstrong R. N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10:2–18. - PubMed
-
- Mannervik B., Danielson U. H. Glutathione transferases: structure and catalytic activity. Crit. Rev. Biochem. Mol. Biol. 1988;23:283–337. - PubMed
-
- Hayes J. D., Flanagan J. U., Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

