Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro
- PMID: 16251280
- PMCID: PMC1280145
- DOI: 10.1128/AAC.49.11.4437-4442.2005
Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro
Abstract
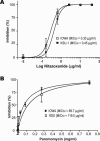
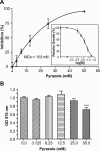
We report here on a quantitative real-time reverse transcription-PCR (qRT-PCR) assay for assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum. The qRT-PCR assay detects 18S rRNA transcripts from both parasites, that is, the cycle threshold for 18S rRNA from parasites (C(T)([P18S])) and host cells (C(T)([H18S])), and evaluates the relative expression between parasite and host rRNA levels (i.e., deltaC(T) = C(T)([P18S]) - C(T)([H18S])) to minimize experimental and operational errors. The choice of qRT-PCR over quantitative PCR (qPCR) in this study is based on the observations that (i) the relationship between the logarithm of infected parasites (log[P]) and the normalized relative level of rRNA (deltadeltaC(T)) is linear, with a fourfold dynamic range, by qRT-PCR but sigmoidal (nonlinear) by qPCR; and (ii) the level of RNA represents that of live parasites better than that of DNA, because the decay of RNA (99% in approximately 3 h) in dead parasites is faster than that of DNA (99% in approximately 24 to 48 h) under in vitro conditions. The reliability of the qRT-PCR method was validated by testing the efficacies of nitazoxanide and paromomycin on the development of two strains of C. parvum (IOWA and KSU-1) in HCT-8 cells in vitro. Both compounds displayed dose-dependent inhibitions. The observed MIC50 values for nitazoxanide and paromomycin were 0.30 to 0.45 micro/ml and 89.7 to 119.0 microg/ml, respectively, comparable to the values reported previously. Using the qRT-PCR assay, we have also observed that pyrazole could inhibit C. parvum development in vitro (MIC50 = 15.8 mM), suggesting that the recently discovered Cryptosporidium alcohol dehydrogenases may be explored as new drug targets.
Figures


References
-
- Abrahamsen, M. S., and A. A. Schroeder. 1999. Characterization of intracellular Cryptosporidium parvum gene expression. Mol. Biochem. Parasitol. 104:141-146. - PubMed
-
- Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. - PubMed
-
- Andersson, P., J. Kvassman, A. Lindstrom, B. Olden, and G. Pettersson. 1981. Effect of pH on pyrazole binding to liver alcohol dehydrogenase. Eur. J. Biochem. 114:549-554. - PubMed
-
- Cai, X., C. A. Lancto, M. S. Abrahamsen, and G. Zhu. 2004. Intron-containing beta-tubulin transcripts in Cryptosporidium parvum cultured in vitro. Microbiology 150:1191-1195. - PubMed
-
- Chappell, C. L., and P. C. Okhuysen. 2002. Cryptosporidiosis. Curr. Opin. Infect. Dis. 15:523-527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

