Mechanisms of azole resistance in a clinical isolate of Candida tropicalis
- PMID: 16251302
- PMCID: PMC1280149
- DOI: 10.1128/AAC.49.11.4608-4615.2005
Mechanisms of azole resistance in a clinical isolate of Candida tropicalis
Abstract
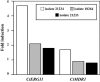
Azole resistance has been insufficiently investigated in the yeast Candida tropicalis. Here we determined the molecular mechanisms responsible for azole resistance in a clinical isolate of this pathogenic yeast. Antifungal susceptibility testing performed by a disk diffusion method showed resistance or markedly decreased susceptibility to azoles, which was confirmed by determination of MICs. Considering the relationship between azole susceptibility and the respiration reported for other yeast species, the respiratory activity of this isolate was investigated. Flow cytometry using rhodamine 123 and oxygraphy demonstrated an increased respiratory activity, which was not linked to an overexpression or increased number of copies of the mitochondrial genome. Among previously described resistance mechanisms, an increased activity of efflux pumps was investigated by flow cytometry using rhodamine 6G. However, the efflux of rhodamine 6G was lower in the resistant isolate than in susceptible ones. Likewise, real-time reverse transcription-PCR quantification of the expression of C. tropicalis MDR1 (CtMDR1), which encodes an efflux protein belonging to the major facilitator superfamily, did not show overexpression of this gene. In contrast, the resistant isolate overexpressed the CtERG11 gene coding for lanosterol 14alpha-demethylase. This was in agreement with the larger amount of ergosterol found in this isolate. Moreover, sequencing of CtERG11 showed a point mutation leading to a tyrosine substitution in the protein sequence, which might lead to decreased binding affinity for azoles. In conclusion, overexpression of CtERG11 associated with a missense mutation in this gene seemed to be responsible for the acquired azole resistance of this clinical isolate.
Figures


References
-
- Casalinuovo, I. A., P. Di Francesco, and E. Garaci. 2004. Fluconazole resistance in Candida albicans: a review of mechanisms. Eur. Rev. Med. Pharmacol. Sci. 8:69-77. - PubMed
-
- Colombo, A. L., and T. Guimaraes. 2003. Epidemiology of hematogenous infections due to Candida spp. Rev. Soc. Bras. Med. Trop. 36:599-607. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

