The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus
- PMID: 16251352
- PMCID: PMC1345684
- DOI: 10.1091/mbc.e05-07-0698
The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus
Abstract
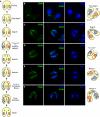
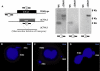
Ciliated protozoans present several features of chromosome segregation that are unique among eukaryotes, including their maintenance of two nuclei: a germline micronucleus, which undergoes conventional mitosis and meiosis, and a somatic macronucleus that divides by an amitotic process. To study ciliate chromosome segregation, we have identified the centromeric histone gene in the Tetrahymena thermophila genome (CNA1). CNA1p specifically localizes to peripheral centromeres in the micronucleus but is absent in the macronucleus during vegetative growth. During meiotic prophase of the micronucleus, when chromosomes are stretched to twice the length of the cell, CNA1p is found localized in punctate spots throughout the length of the chromosomes. As conjugation proceeds, CNA1p appears initially diffuse, but quickly reverts to discrete dots in those nuclei destined to become micronuclei, whereas it remains diffuse and is gradually lost in developing macronuclei. In progeny of germline CNA1 knockouts, we see no defects in macronuclear division or viability of the progeny cells immediately following the knockout. However, within a few divisions, progeny show abnormal mitotic segregation of their micronucleus, with most cells eventually losing their micronucleus entirely. This study reveals a strong dependence of the germline micronucleus on centromeric histones for proper chromosome segregation.
Figures





Similar articles
-
Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila.Mol Cell Biol. 2006 Jun;26(12):4499-510. doi: 10.1128/MCB.00079-06. Mol Cell Biol. 2006. PMID: 16738316 Free PMC article.
-
Phosphorylation of histone H3 is required for proper chromosome condensation and segregation.Cell. 1999 Apr 2;97(1):99-109. doi: 10.1016/s0092-8674(00)80718-7. Cell. 1999. PMID: 10199406
-
Kinesin-14 is Important for Chromosome Segregation During Mitosis and Meiosis in the Ciliate Tetrahymena thermophila.J Eukaryot Microbiol. 2017 May;64(3):293-307. doi: 10.1111/jeu.12366. Epub 2016 Sep 23. J Eukaryot Microbiol. 2017. PMID: 27595611
-
Developmental progression of Tetrahymena through the cell cycle and conjugation.Methods Cell Biol. 2012;109:177-236. doi: 10.1016/B978-0-12-385967-9.00007-4. Methods Cell Biol. 2012. PMID: 22444146 Review.
-
Dynamic nuclear reorganization during genome remodeling of Tetrahymena.Biochim Biophys Acta. 2008 Nov;1783(11):2130-6. doi: 10.1016/j.bbamcr.2008.07.012. Epub 2008 Jul 28. Biochim Biophys Acta. 2008. PMID: 18706458 Free PMC article. Review.
Cited by
-
Cyc17, a meiosis-specific cyclin, is essential for anaphase initiation and chromosome segregation in Tetrahymena thermophila.Cell Cycle. 2016 Jul 17;15(14):1855-64. doi: 10.1080/15384101.2016.1188238. Epub 2016 May 18. Cell Cycle. 2016. PMID: 27192402 Free PMC article.
-
The histone chaperone Nrp1 is required for chromatin stability and nuclear division in Tetrahymena thermophila.Epigenetics Chromatin. 2021 Jul 23;14(1):34. doi: 10.1186/s13072-021-00409-4. Epigenetics Chromatin. 2021. PMID: 34301312 Free PMC article.
-
Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes.Res Microbiol. 2011 Jul-Aug;162(6):578-86. doi: 10.1016/j.resmic.2011.05.001. Epub 2011 May 18. Res Microbiol. 2011. PMID: 21624459 Free PMC article. Review.
-
Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila.Mol Cell Biol. 2006 Jun;26(12):4499-510. doi: 10.1128/MCB.00079-06. Mol Cell Biol. 2006. PMID: 16738316 Free PMC article.
-
Exploration of the Nuclear Proteomes in the Ciliate Oxytricha trifallax.Microorganisms. 2023 Jan 30;11(2):343. doi: 10.3390/microorganisms11020343. Microorganisms. 2023. PMID: 36838311 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

