DNA replication origins fire stochastically in fission yeast
- PMID: 16251353
- PMCID: PMC1345668
- DOI: 10.1091/mbc.e05-07-0657
DNA replication origins fire stochastically in fission yeast
Abstract
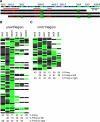
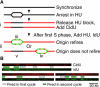
DNA replication initiates at discrete origins along eukaryotic chromosomes. However, in most organisms, origin firing is not efficient; a specific origin will fire in some but not all cell cycles. This observation raises the question of how individual origins are selected to fire and whether origin firing is globally coordinated to ensure an even distribution of replication initiation across the genome. We have addressed these questions by determining the location of firing origins on individual fission yeast DNA molecules using DNA combing. We show that the firing of replication origins is stochastic, leading to a random distribution of replication initiation. Furthermore, origin firing is independent between cell cycles; there is no epigenetic mechanism causing an origin that fires in one cell cycle to preferentially fire in the next. Thus, the fission yeast strategy for the initiation of replication is different from models of eukaryotic replication that propose coordinated origin firing.
Figures

Similar articles
-
Checkpoint independence of most DNA replication origins in fission yeast.BMC Mol Biol. 2007 Dec 19;8:112. doi: 10.1186/1471-2199-8-112. BMC Mol Biol. 2007. PMID: 18093330 Free PMC article.
-
DNA replication in the fission yeast: robustness in the face of uncertainty.Yeast. 2006 Oct 15;23(13):951-62. doi: 10.1002/yea.1416. Yeast. 2006. PMID: 17072888 Review.
-
Molecular Combing of Single DNA Molecules on the 10 Megabase Scale.Sci Rep. 2016 Jan 19;6:19636. doi: 10.1038/srep19636. Sci Rep. 2016. PMID: 26781994 Free PMC article.
-
Specific replication origins promote DNA amplification in fission yeast.J Cell Sci. 2010 Sep 15;123(Pt 18):3047-51. doi: 10.1242/jcs.067470. Epub 2010 Aug 24. J Cell Sci. 2010. PMID: 20736315 Free PMC article.
-
Surveying genome replication.Genome Biol. 2002;3(6):REVIEWS1016. doi: 10.1186/gb-2002-3-6-reviews1016. Epub 2002 May 24. Genome Biol. 2002. PMID: 12093380 Free PMC article. Review.
Cited by
-
GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris.PLoS Genet. 2014 Mar 6;10(3):e1004169. doi: 10.1371/journal.pgen.1004169. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603708 Free PMC article.
-
[Regulation of DNA replication timing].Mol Biol (Mosk). 2013 Jan-Feb;47(1):12-37. doi: 10.1134/s0026893312060118. Mol Biol (Mosk). 2013. PMID: 23705493 Review. Russian.
-
Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers.Proc Natl Acad Sci U S A. 2016 Aug 16;113(33):E4810-9. doi: 10.1073/pnas.1609060113. Epub 2016 Jul 19. Proc Natl Acad Sci U S A. 2016. PMID: 27436900 Free PMC article.
-
Genome-wide identification and characterization of replication origins by deep sequencing.Genome Biol. 2012 Apr 24;13(4):R27. doi: 10.1186/gb-2012-13-4-r27. Genome Biol. 2012. PMID: 22531001 Free PMC article.
-
Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast.EMBO J. 2012 Jan 4;31(1):124-37. doi: 10.1038/emboj.2011.350. Epub 2011 Oct 11. EMBO J. 2012. PMID: 21989386 Free PMC article.
References
-
- Abdurashidova, G., Deganuto, M., Klima, R., Riva, S., Biamonti, G., Giacca, M., and Falaschi, A. (2000). Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287, 2023-2026. - PubMed
-
- Anglana, M., Apiou, F., Bensimon, A., and Debatisse, M. (2003). Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114, 385-394. - PubMed
-
- Bensimon, A., Simon, A., Chiffaudel, A., Croquette, V., Heslot, F., and Bensimon, D. (1994). Alignment and sensitive detection of DNA by a moving interface. Science 265, 2096-2098. - PubMed
-
- Berezney, R., Dubey, D. D., and Huberman, J. A. (2000). Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108, 471-484. - PubMed
-
- Bielinsky, A. K., and Gerbi, S. A. (1999). Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3, 477-486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

