Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
- PMID: 16251366
- PMCID: PMC2253226
- DOI: 10.1110/ps.051666705
Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Abstract
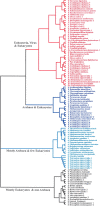
The solution structure of protein AF2095 from the thermophilic archaea Archaeglobus fulgidis, a 123-residue (13.6-kDa) protein, has been determined by NMR methods. The structure of AF2095 is comprised of four alpha-helices and a mixed beta-sheet consisting of four parallel and anti-parallel beta-strands, where the alpha-helices sandwich the beta-sheet. Sequence and structural comparison of AF2095 with proteins from Homo sapiens, Methanocaldococcus jannaschii, and Sulfolobus solfataricus reveals that AF2095 is a peptidyl-tRNA hydrolase (Pth2). This structural comparison also identifies putative catalytic residues and a tRNA interaction region for AF2095. The structure of AF2095 is also similar to the structure of protein TA0108 from archaea Thermoplasma acidophilum, which is deposited in the Protein Data Bank but not functionally annotated. The NMR structure of AF2095 has been further leveraged to obtain good-quality structural models for 55 other proteins. Although earlier studies have proposed that the Pth2 protein family is restricted to archeal and eukaryotic organisms, the similarity of the AF2095 structure to human Pth2, the conservation of key active-site residues, and the good quality of the resulting homology models demonstrate a large family of homologous Pth2 proteins that are conserved in eukaryotic, archaeal, and bacterial organisms, providing novel insights in the evolution of the Pth and Pth2 enzyme families.
Figures
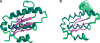


Similar articles
-
Orthologs of a novel archaeal and of the bacterial peptidyl-tRNA hydrolase are nonessential in yeast.Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16707-12. doi: 10.1073/pnas.222659199. Epub 2002 Dec 10. Proc Natl Acad Sci U S A. 2002. PMID: 12475929 Free PMC article.
-
Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity.J Biol Chem. 2004 Feb 27;279(9):8111-5. doi: 10.1074/jbc.M311449200. Epub 2003 Dec 5. J Biol Chem. 2004. PMID: 14660562
-
Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.Biochemistry. 2005 Mar 22;44(11):4294-301. doi: 10.1021/bi047711k. Biochemistry. 2005. PMID: 15766258
-
Structural and functional insights into peptidyl-tRNA hydrolase.Biochim Biophys Acta. 2014 Jul;1844(7):1279-88. doi: 10.1016/j.bbapap.2014.04.012. Epub 2014 Apr 21. Biochim Biophys Acta. 2014. PMID: 24768774 Review.
-
RNase P: interface of the RNA and protein worlds.Trends Biochem Sci. 2006 Jun;31(6):333-41. doi: 10.1016/j.tibs.2006.04.007. Epub 2006 May 6. Trends Biochem Sci. 2006. PMID: 16679018 Review.
Cited by
-
Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Å resolution shows the closed structure of the substrate-binding cleft.FEBS Open Bio. 2014 Oct 22;4:915-22. doi: 10.1016/j.fob.2014.10.010. eCollection 2014. FEBS Open Bio. 2014. PMID: 25389518 Free PMC article.
-
Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex.Proteins. 2009 Apr;75(1):12-27. doi: 10.1002/prot.22217. Proteins. 2009. PMID: 18767154 Free PMC article.
-
Neutron diffraction analysis of Pseudomonas aeruginosa peptidyl-tRNA hydrolase 1.Acta Crystallogr F Struct Biol Commun. 2016 Mar;72(Pt 3):220-3. doi: 10.1107/S2053230X16001813. Epub 2016 Feb 19. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 26919526 Free PMC article.
-
Small Molecule Docking Supports Broad and Narrow Spectrum Potential for the Inhibition of the Novel Antibiotic Target Bacterial Pth1.Antibiotics (Basel). 2016 May 10;5(2):16. doi: 10.3390/antibiotics5020016. Antibiotics (Basel). 2016. PMID: 27171117 Free PMC article.
-
Identification of a Ligand-Binding Site on the Staphylococcus aureus DnaG Primase C-Terminal Domain.Biochemistry. 2017 Feb 21;56(7):932-943. doi: 10.1021/acs.biochem.6b01273. Epub 2017 Feb 9. Biochemistry. 2017. PMID: 28125218 Free PMC article.
References
-
- Acton, T.B., Gunsalus, K.C., Xiao, R., Ma, L.C., Aramini, J., Baran, M.C., Chiang, Y.-W., Climent, T., Cooper, B., Denissova, N.G., et al. 2005. Robotic cloning and protein production platform of the northeast structural genomics consortium. Methods Enzymol. 394 210–243. - PubMed
-
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4 187–217.
-
- Brun, G., Paulin, D., Yot, P., and Chapeville, F. 1971. Peptidyl-tRNA hydrolase: Characterization in some organisms. Enzymic activity in the presence of ribosomes. Biochimie 53 225–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

