Predicting RNA folding thermodynamics with a reduced chain representation model
- PMID: 16251382
- PMCID: PMC1370876
- DOI: 10.1261/rna.2109105
Predicting RNA folding thermodynamics with a reduced chain representation model
Abstract
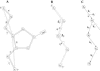
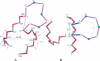
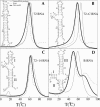
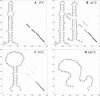
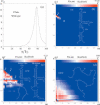
Based on the virtual bond representation for the nucleotide backbone, we develop a reduced conformational model for RNA. We use the experimentally measured atomic coordinates to model the helices and use the self-avoiding walks in a diamond lattice to model the loop conformations. The atomic coordinates of the helices and the lattice representation for the loops are matched at the loop-helix junction, where steric viability is accounted for. Unlike the previous simplified lattice-based models, the present virtual bond model can account for the atomic details of realistic three-dimensional RNA structures. Based on the model, we develop a statistical mechanical theory for RNA folding energy landscapes and folding thermodynamics. Tests against experiments show that the theory can give much more improved predictions for the native structures, the thermal denaturation curves, and the equilibrium folding/unfolding pathways than the previous models. The application of the model to the P5abc region of Tetrahymena group I ribozyme reveals the misfolded intermediates as well as the native-like intermediates in the equilibrium folding process. Moreover, based on the free energy landscape analysis for each and every loop mutation, the model predicts five lethal mutations that can completely alter the free energy landscape and the folding stability of the molecule.
Figures
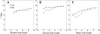
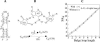




References
-
- Biswas, R., Mitra, S.N., and Sundaralingam, M. 1998. 1.76Å structure of a pyrimidine start alternating A-RNA hexamer r(CGUAC)dG. Acta Cryst. D 54: 570–576. - PubMed
-
- Chan, H.S., Shimizu, S., and Kaya, H. 2004. Cooperativity principles in protein folding. Methods Enzymol. 380: 350–379. - PubMed
-
- Chen, S.-J. and Dill, K.A. 1995. Statistical thermodynamics of doublestranded polymer molecules. J. Chem. Phys. 103: 5802–5813.
-
- ———. 1998. Theory for the conformational changes of doublestranded chain molecules. J. Chem. Phys. 109: 4602–4616.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
