Functional characterization of the SOFA delta ribozyme
- PMID: 16251383
- PMCID: PMC1370874
- DOI: 10.1261/rna.2112705
Functional characterization of the SOFA delta ribozyme
Abstract
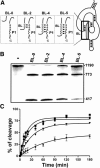
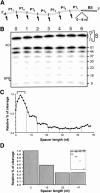
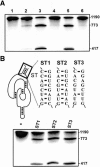
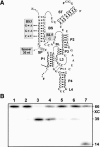
Molecular engineering has led to the development of a novel target-dependent riboswitch that increases deltaribozyme fidelity. This delta ribozyme possesses a specific on/off adapter (SOFA) that switches the cleavage activity from off (a "safety lock") to on solely in the presence of the desired RNA substrate. In this report, we investigate the influence of both the structure and the sequence of each domain of the SOFA module. Analysis of the cleavage activity, using a large collection of substrates and SOFA-ribozyme mutants, together with RNase H probing provided several insights into the nature of the sequence and the optimal design of each domain of the SOFA module. For example, we determined that (1) the optimal size of the blocker sequence, which keeps the ribozyme off in the absence of the substrate, is 4 nucleotides (nt); (2) a single nucleotide difference between the substrate and the biosensor domain, which is responsible for the initial binding of the substrate that subsequently switches the SOFA-ribozyme on, is sufficient to cause non-recognition of the appropriate substrate; (3) the stabilizer, which joins the 5' and 3' ends of the SOFA-ribozyme, plays only a structural role; and (4) the optimal spacer sequence, which serves to separate the binding regions of the biosensor and catalytic domain of the ribozyme on the substrate, is from 1 to 5 nt long. Together, these data should facilitate the design of more efficient SOFA-ribozymes with significant potential for many applications in gene-inactivation systems.
Figures




Similar articles
-
Experimental evidence for the secondary structure of the hepatitis delta virus ribozyme.Prog Clin Biol Res. 1993;382:69-77. Prog Clin Biol Res. 1993. PMID: 8502719
-
Trans cleavage of RNA substrates by an HDV-derived ribozyme.Prog Clin Biol Res. 1993;382:99-108. Prog Clin Biol Res. 1993. PMID: 8502721 No abstract available.
-
Target-induced SOFA-HDV ribozyme.Methods Mol Biol. 2012;848:369-84. doi: 10.1007/978-1-61779-545-9_23. Methods Mol Biol. 2012. PMID: 22315081
-
Ribozymes of the hepatitis delta virus: recent findings on their structure, mechanism of catalysis and possible applications.Acta Biochim Pol. 2001;48(2):409-18. Acta Biochim Pol. 2001. PMID: 11732611 Review.
-
Development of ribozyme-based gene-inactivations; the example of the hepatitis delta virus ribozyme.Curr Gene Ther. 2007 Jun;7(3):205-16. doi: 10.2174/156652307780859008. Curr Gene Ther. 2007. PMID: 17584038 Review.
Cited by
-
A modern mode of activation for nucleic acid enzymes.PLoS One. 2007 Jul 25;2(7):e673. doi: 10.1371/journal.pone.0000673. PLoS One. 2007. PMID: 17653287 Free PMC article.
-
Effective inhibition of HIV-1 production by short hairpin RNAs and small interfering RNAs targeting a highly conserved site in HIV-1 Gag RNA is optimized by evaluating alternative length formats.Antimicrob Agents Chemother. 2015 Sep;59(9):5297-305. doi: 10.1128/AAC.00949-15. Epub 2015 Jun 15. Antimicrob Agents Chemother. 2015. PMID: 26077260 Free PMC article.
-
Topological constraints of structural elements in regulation of catalytic activity in HDV-like self-cleaving ribozymes.Sci Rep. 2016 Jun 15;6:28179. doi: 10.1038/srep28179. Sci Rep. 2016. PMID: 27302490 Free PMC article.
-
Gene targeting in the Gram-Positive bacterium Lactococcus lactis, using various delta ribozymes.Appl Environ Microbiol. 2006 Jan;72(1):869-79. doi: 10.1128/AEM.72.1.869-879.2006. Appl Environ Microbiol. 2006. PMID: 16391129 Free PMC article.
-
Prospects for nucleic acid-based therapeutics against hepatitis C virus.World J Gastroenterol. 2013 Dec 21;19(47):8949-62. doi: 10.3748/wjg.v19.i47.8949. World J Gastroenterol. 2013. PMID: 24379620 Free PMC article. Review.
References
-
- Al-Anouti, F. and Ananvoranich, S. 2002. Comparative analysis of antisense RNA, double stranded RNA, and delta ribozymemediated gene regulation in toxoplasma gondii. Antisense Nucleic Acid Drug Dev. 12: 275–281. - PubMed
-
- Bagheri, S. and Kashani-Sabet, M. 2004. Ribozymes in the age of molecular therapeutics. Curr. Mol. Med. 4: 489–506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
