Genomic regions exhibiting positive selection identified from dense genotype data
- PMID: 16251465
- PMCID: PMC1310643
- DOI: 10.1101/gr.4326505
Genomic regions exhibiting positive selection identified from dense genotype data
Abstract
The allele frequency spectrum of polymorphisms in DNA sequences can be used to test for signatures of natural selection that depart from the expected frequency spectrum under the neutral theory. We observed a significant (P = 0.001) correlation between the Tajima's D test statistic in full resequencing data and Tajima's D in a dense, genome-wide data set of genotyped polymorphisms for a set of 179 genes. Based on this, we used a sliding window analysis of Tajima's D across the human genome to identify regions putatively subject to strong, recent, selective sweeps. This survey identified seven Contiguous Regions of Tajima's D Reduction (CRTRs) in an African-descent population (AD), 23 in a European-descent population (ED), and 29 in a Chinese-descent population (XD). Only four CRTRs overlapped between populations: three between ED and XD and one between AD and ED. Full resequencing of eight genes within six CRTRs demonstrated frequency spectra inconsistent with neutral expectations for at least one gene within each CRTR. Identification of the functional polymorphism (and/or haplotype) responsible for the selective sweeps within each CRTR may provide interesting insights into the strongest selective pressures experienced by the human genome over recent evolutionary history.
Figures

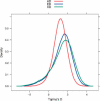


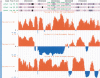
References
-
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518–521. - PubMed
-
- Clark, A.G., Glanowski, S., Nielsen, R., Thomas, P., Kejariwal, A., Todd, M.J., Tanenbaum, D.M., Civello, D., Lu, F., Murphy, B., et al. 2003a. Positive selection in the human genome inferred from human–chimp–mouse orthologous gene alignments. Cold Spring Harb. Symp. Quant. Biol. 68: 471–477. - PubMed
Web site references
-
- http://pga.gs.washington.edu; Seattle SNPs Web site.
-
- http://genome.perlegen.com/browser/download.html; Perlegen Web site.
-
- http://genome.ucsc.edu/cgi-bin/hgGateway; UCSC Genome Browser.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
