Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
- PMID: 16252003
- PMCID: PMC1356300
- DOI: 10.1038/sj.emboj.7600858
Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Abstract
Pleckstrin homology (PH) domains play diverse roles in cytoskeletal dynamics and signal transduction. Split PH domains represent a unique subclass of PH domains that have been implicated in interactions with complementary partial PH domains 'hidden' in many proteins. Whether partial PH domains exist as independent structural units alone and whether two halves of a split PH domain can fold together to form an intact PH domain are not known. Here, we solved the structure of the PH(N)-PDZ-PH(C) tandem of alpha-syntrophin. The split PH domain of alpha-syntrophin adopts a canonical PH domain fold. The isolated partial PH domains of alpha-syntrophin, although completely unfolded, remain soluble in solution. Mixing of the two isolated domains induces de novo folding and yields a stable PH domain. Our results demonstrate that two complementary partial PH domains are capable of binding to each other to form an intact PH domain. We further showed that the PH(N)-PDZ-PH(C) tandem forms a functionally distinct supramodule, in which the split PH domain and the PDZ domain function synergistically in binding to inositol phospholipids.
Figures
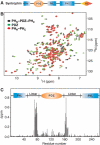
 ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts between the PHN–PDZ–PHC tandem and the isolated PH and PDZ domains, respectively. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17. The domain organization of the PHN–PDZ–PHC tandem is indicated at the top of the plot.
ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts between the PHN–PDZ–PHC tandem and the isolated PH and PDZ domains, respectively. The scaling factor (αN) used to normalize the 1H and 15N chemical shifts is 0.17. The domain organization of the PHN–PDZ–PHC tandem is indicated at the top of the plot.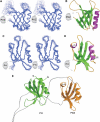



References
-
- Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, Froehner SC (1993) Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron 11: 531–540 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

