Promoter and 3'-untranslated-region haplotypes in the vitamin d receptor gene predispose to osteoporotic fracture: the rotterdam study
- PMID: 16252240
- PMCID: PMC1271389
- DOI: 10.1086/497438
Promoter and 3'-untranslated-region haplotypes in the vitamin d receptor gene predispose to osteoporotic fracture: the rotterdam study
Abstract
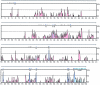
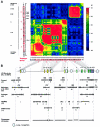
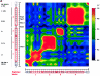
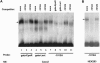
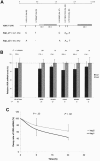
Polymorphisms of the vitamin D receptor gene (VDR) have been shown to be associated with several complex diseases, including osteoporosis, but the mechanisms are unknown and study results have been inconsistent. We therefore determined sequence variation across the major relevant parts of VDR, including construction of linkage disequilibrium blocks and identification of haplotype alleles. We analyzed 15 haplotype-tagging SNPs in relation to 937 clinical fractures recorded in 6,148 elderly whites over a follow-up period of 7.4 years. Haplotype alleles of the 5' 1a/1e, 1b promoter region and of the 3' untranslated region (UTR) were strongly associated with increased fracture risk. For the 16% of subjects who had risk genotypes at both regions, their risk increased 48% for clinical fractures (P = .0002), independent of age, sex, height, weight, and bone mineral density. The population-attributable risk varied between 1% and 12% for each block and was 4% for the combined VDR risk genotypes. Functional analysis of the variants demonstrated 53% lower expression of a reporter construct with the 1e/1a promoter risk haplotype (P = 5 x 10(-7)) in two cell lines and 15% lower mRNA level of VDR expression constructs carrying 3'-UTR risk haplotype 1 in five cell lines (P = 2 x 10(-6)). In a further analysis, we showed 30% increased mRNA decay in an osteoblast cell line for the construct carrying the 3'-UTR risk haplotype (P = .02). This comprehensive candidate-gene analysis demonstrates that the risk allele of multiple VDR polymorphisms results in lower VDR mRNA levels. This could impact the vitamin D signaling efficiency and might contribute to the increased fracture risk we observed for these risk haplotype alleles.
Figures







References
Web Resources
-
- Applied Biosystems SNPbrowser Software, http://events-na.appliedbiosystems.com/mk/get/snpb_landing?isource=fr_E_...
-
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for detected polymorphisms [accession numbers ss32465817–ss32465820 and ss32465633–ss32465690])
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for reference sequences [accession numbers BV210413–BV210452 and AY827085–AY827088])
References
-
- Arai H, Miyamoto KI, Yoshida M, Yamamoto H, Taketani Y, Morita K, Kubota M, Yoshida S, Ikeda M, Watabe F, Kanemasa Y, Takeda E (2001) The polymorphism in the caudal-related homeodomain protein Cdx-2 binding element in the human vitamin D receptor gene. J Bone Miner Res 16:1256–1264 - PubMed
-
- Brown SB, Brierley TT, Palanisamy N, Salusky IB, Goodman W, Brandi ML, Drueke TB, Sarfati E, Urena P, Chaganti RS, Pike JW, Arnold A (2000) Vitamin D receptor as a candidate tumor-suppressor gene in severe hyperparathyroidism of uremia. J Clin Endocrinol Metab 85:868–872 - PubMed
-
- Burger H, van Daele PL, Algra D, van den Ouweland FA, Grobbee DE, Hofman A, van Kuijk C, Schutte HE, Birkenhager JC, Pols HA (1994) The association between age and bone mineral density in men and women aged 55 years and over: the Rotterdam Study. Bone Miner 25:1–13 - PubMed
-
- Carling T, Ridefelt P, Hellman P, Rastad J, Akerstrom G (1997) Vitamin D receptor polymorphisms correlate to parathyroid cell function in primary hyperparathyroidism. J Clin Endocrinol Metab 82:1772–1775 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

