Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus
- PMID: 16252243
- PMCID: PMC1271392
- DOI: 10.1086/497705
Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus
Abstract
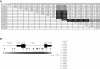
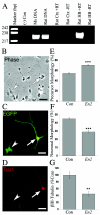
Our previous research involving 167 nuclear families from the Autism Genetic Resource Exchange (AGRE) demonstrated that two intronic SNPs, rs1861972 and rs1861973, in the homeodomain transcription factor gene ENGRAILED 2 (EN2) are significantly associated with autism spectrum disorder (ASD). In this study, significant replication of association for rs1861972 and rs1861973 is reported for two additional data sets: an independent set of 222 AGRE families (rs1861972-rs1861973 haplotype, P=.0016) and a separate sample of 129 National Institutes of Mental Health families (rs1861972-rs1861973 haplotype, P=.0431). Association analysis of the haplotype in the combined sample of both AGRE data sets (389 families) produced a P value of .0000033, whereas combining all three data sets (518 families) produced a P value of .00000035. Population-attributable risk calculations for the associated haplotype, performed using the entire sample of 518 families, determined that the risk allele contributes to as many as 40% of ASD cases in the general population. Linkage disequilibrium (LD) mapping with the use of polymorphisms distributed throughout the gene has shown that only intronic SNPs are in strong LD with rs1861972 and rs1861973. Resequencing and association analysis of all intronic SNPs have identified alleles associated with ASD, which makes them candidates for future functional analysis. Finally, to begin defining the function of EN2 during development, mouse En2 was ectopically expressed in cortical precursors. Fewer En2-transfected cells than controls displayed a differentiated phenotype. Together, these data provide further genetic evidence that EN2 might act as an ASD susceptibility locus, and they suggest that a risk allele that perturbs the spatial/temporal expression of EN2 could significantly alter normal brain development.
Figures

References
Web Resources
-
- Autism Genetic Resource Exchange (AGRE), http://www.agre.org/
-
- National Institute of Mental Health (NIMH) Data Set, http://zork.wustl.edu/nimh/NIMH_initiative/NIMH_initiative_link.html
-
- NIMH Center for Collaborative Genetic Studies, http://zork.wustl.edu/nimh/home/d_autism.html
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ASD and EN2) - PubMed
References
-
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 - PubMed
-
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J (2000) Single nucleotide polymorphism analysis by pyrosequencing. Anal Biochem 280:103–110 - PubMed
-
- Akshoomoff NA, Courchesne E, Townsend J (1997) Attention coordination and anticipatory control. Int Rev Neurobiol 41:575–598 - PubMed
-
- Allen G, Buxton RB, Wong EC, Courchesne E (1997) Attentional activation of the cerebellum independent of motor involvement. Science 275:1940–1943 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH70366/MH/NIMH NIH HHS/United States
- P30 ES005022/ES/NIEHS NIH HHS/United States
- MH52708/MH/NIMH NIH HHS/United States
- P01 ES11256/ES/NIEHS NIH HHS/United States
- 1 F30 NS48649-01/NS/NINDS NIH HHS/United States
- P01 ES011256/ES/NIEHS NIH HHS/United States
- MH00219/MH/NIMH NIH HHS/United States
- F30 NS048649/NS/NINDS NIH HHS/United States
- R01 MH070366/MH/NIMH NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- T32 MH019957/MH/NIMH NIH HHS/United States
- MH39437/MH/NIMH NIH HHS/United States
- MH068457/MH/NIMH NIH HHS/United States
- MH/AG-19957/MH/NIMH NIH HHS/United States
- MH00980/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

