The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region
- PMID: 16252244
- PMCID: PMC1271393
- DOI: 10.1086/497613
The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region
Abstract
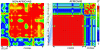
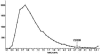
The innate immunity system constitutes the first line of host defense against pathogens. Two closely related innate immunity genes, CD209 and CD209L, are particularly interesting because they directly recognize a plethora of pathogens, including bacteria, viruses, and parasites. Both genes, which result from an ancient duplication, possess a neck region, made up of seven repeats of 23 amino acids each, known to play a major role in the pathogen-binding properties of these proteins. To explore the extent to which pathogens have exerted selective pressures on these innate immunity genes, we resequenced them in a group of samples from sub-Saharan Africa, Europe, and East Asia. Moreover, variation in the number of repeats of the neck region was defined in the entire Human Genome Diversity Panel for both genes. Our results, which are based on diversity levels, neutrality tests, population genetic distances, and neck-region length variation, provide genetic evidence that CD209 has been under a strong selective constraint that prevents accumulation of any amino acid changes, whereas CD209L variability has most likely been shaped by the action of balancing selection in non-African populations. In addition, our data point to the neck region as the functional target of such selective pressures: CD209 presents a constant size in the neck region populationwide, whereas CD209L presents an excess of length variation, particularly in non-African populations. An additional interesting observation came from the coalescent-based CD209 gene tree, whose binary topology and time depth (approximately 2.8 million years ago) are compatible with an ancestral population structure in Africa. Altogether, our study has revealed that even a short segment of the human genome can uncover an extraordinarily complex evolutionary history, including different pathogen pressures on host genes as well as traces of admixture among archaic hominid populations.
Figures
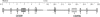




References
Web Resources
-
- Arlequin, http://lgb.unige.ch/arlequin/
-
- Center for Statistical Genetics, http://www.sph.umich.edu/csg/abecasis/GOLD/ (for GOLD software)
-
- Centre National de Genotypage, http://software.cng.fr/ (for GENALYS software)
-
- DnaSP, http://www.ub.es/dnasp/
References
-
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 - PubMed
-
- Appelmelk BJ, van Die I, van Vliet SJ, Vandenbroucke-Grauls CM, Geijtenbeek TB, van Kooyk Y (2003) Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J Immunol 170:1635–1639 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

