Induction of unique structural changes in guanine-rich DNA regions by the triazoloacridone C-1305, a topoisomerase II inhibitor with antitumor activities
- PMID: 16254080
- PMCID: PMC1270948
- DOI: 10.1093/nar/gki904
Induction of unique structural changes in guanine-rich DNA regions by the triazoloacridone C-1305, a topoisomerase II inhibitor with antitumor activities
Abstract
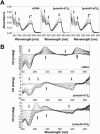
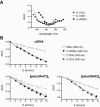
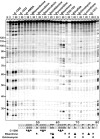
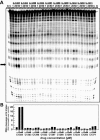
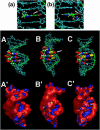
We recently reported that the antitumor triazoloacridone, compound C-1305, is a topoisomerase II poison with unusual properties. In this study we characterize the DNA interactions of C-1305 in vitro, in comparison with other topoisomerase II inhibitors. Our results show that C-1305 binds to DNA by intercalation and possesses higher affinity for GC- than AT-DNA as revealed by surface plasmon resonance studies. Chemical probing with DEPC indicated that C-1305 induces structural perturbations in DNA regions with three adjacent guanine residues. Importantly, this effect was highly specific for C-1305 since none of the other 22 DNA interacting drugs tested was able to induce similar structural changes in DNA. Compound C-1305 induced stronger structural changes in guanine triplets at higher pH which suggested that protonation/deprotonation of the drug is important for this drug-specific effect. Molecular modeling analysis predicts that the zwitterionic form of C-1305 intercalates within the guanine triplet, resulting in widening of both DNA grooves and aligning of the triazole ring with the N7 atoms of guanines. Our results show that C-1305 binds to DNA and induces very specific and unusual structural changes in guanine triplets which likely plays an important role in the cytotoxic and antitumor activity of this unique compound.
Figures





Similar articles
-
Crystal structure of the topoisomerase II poison 9-amino-[N-(2-dimethylamino)ethyl]acridine-4-carboxamide bound to the DNA hexanucleotide d(CGTACG)2.Biochemistry. 1999 Jul 20;38(29):9221-33. doi: 10.1021/bi990352m. Biochemistry. 1999. PMID: 10413496
-
Kinetic studies of the binding of acridinecarboxamide topoisomerase poisons to DNA: implications for mode of binding of ligands with uncharged chromophores.J Med Chem. 2002 Feb 14;45(4):894-901. doi: 10.1021/jm000473g. J Med Chem. 2002. PMID: 11831901
-
AK37: the first pyridoacridine described capable of stabilizing the topoisomerase I cleavable complex.Anticancer Drugs. 2004 Oct;15(9):907-13. doi: 10.1097/00001813-200410000-00012. Anticancer Drugs. 2004. PMID: 15457132
-
Crystal structures of acridines complexed with nucleic acids.Curr Med Chem. 2002 Sep;9(18):1667-75. doi: 10.2174/0929867023369259. Curr Med Chem. 2002. PMID: 12171549 Review.
-
A new look at 9-substituted acridines with various biological activities.J Appl Toxicol. 2021 Jan;41(1):175-189. doi: 10.1002/jat.4072. Epub 2020 Sep 24. J Appl Toxicol. 2021. PMID: 32969520 Review.
Cited by
-
Electrochemical and in silico approaches for liver metabolic oxidation of antitumor-active triazoloacridinone C-1305.J Pharm Anal. 2020 Aug;10(4):376-384. doi: 10.1016/j.jpha.2020.03.011. Epub 2020 Mar 23. J Pharm Anal. 2020. PMID: 32923012 Free PMC article.
-
Utilizing Genome-Wide mRNA Profiling to Identify the Cytotoxic Chemotherapeutic Mechanism of Triazoloacridone C-1305 as Direct Microtubule Stabilization.Cancers (Basel). 2020 Apr 2;12(4):864. doi: 10.3390/cancers12040864. Cancers (Basel). 2020. PMID: 32252403 Free PMC article.
-
Synthesis, Stability, and Biological Evaluation of Novel Aminoderivatives Incorporating the Aza-Acridine Scaffold.Molecules. 2025 Jun 16;30(12):2612. doi: 10.3390/molecules30122612. Molecules. 2025. PMID: 40572575 Free PMC article.
-
Dual-Action Therapeutics: DNA Alkylation and Antimicrobial Peptides for Cancer Therapy.Cancers (Basel). 2024 Sep 10;16(18):3123. doi: 10.3390/cancers16183123. Cancers (Basel). 2024. PMID: 39335095 Free PMC article. Review.
-
Effective Drug Concentration and Selectivity Depends on Fraction of Primitive Cells.Int J Mol Sci. 2021 May 6;22(9):4931. doi: 10.3390/ijms22094931. Int J Mol Sci. 2021. PMID: 34066491 Free PMC article.
References
-
- Cholody W.M., Martelli S., Konopa J. 8-Substituted 5-[(aminoalkyl)amino]-6H-v-triazolo[4,5,1-de]acridin-6-ones as potential antineoplastic agents. Synthesis and biological activity. J. Med. Chem. 1990;33:2852–2856. - PubMed
-
- Kusnierczyk H., Cholody W.M., Paradziej-Lukowicz J., Radzikowski C., Konopa J. Experimental antitumor activity and toxicity of the selected triazolo- and imidazoacridinones. Arch. Immunol. Ther. Exp. (Warsz.) 1994;42:415–423. - PubMed
-
- Lemke K., Poindessous V., Skladanowski A., Larsen A.K. The antitumor triazoloacridone C-1305 is a topoisomerase II poison with unusual properties. Mol. Pharmacol. 2004;66:1035–1042. - PubMed
-
- Wesierska-Gadek J., Schloffer D., Gueorguieva M., Uhl M., Skladanowski A. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout cells to antitumor triazoloacridone C-1305 is associated with permanent G2 cell cycle arrest. Cancer Res. 2004;64:4487–4497. - PubMed
-
- Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell. Biol. 2002;3:430–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

