Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly
- PMID: 16254319
- PMCID: PMC1280203
- DOI: 10.1128/JVI.79.22.13839-13847.2005
Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly
Abstract
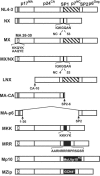
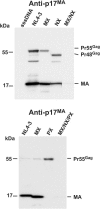
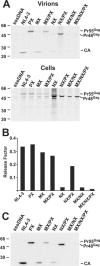
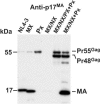
RNA appears to be required for the assembly of retroviruses. This is likely due to binding of RNA by multiple Gags, which in turn organizes and stabilizes the Gag-Gag interactions that form the virion. While the nucleocapsid (NC) domain is the most conspicuous RNA-binding region of the human immunodeficiency virus type 1 (HIV-1) Gag polyprotein, we have previously shown that NC is not strictly required for efficient particle production. To determine if an RNA requirement for HIV-1 assembly exists, we analyzed virions produced by an NC deletion mutant for the presence of RNA. The results revealed that virions without NC still contained significant amounts of RNA. Since these packaged RNAs are probably incorporated by other RNA-binding sequences in Gag, an RNA-binding site in the matrix protein (MA) of Gag was mutated. While this mutation did not interfere with HIV-1 replication, a construct with both MA and NC mutations (MX/NX) failed to produce particles. The MX/NX mutant was rescued in trans by coassembly with several forms of Gag: wild-type Gag, either of the single-mutant Gags, or Gag truncations that contain MA or NC sequences. Addition of basic sequences to the MX/NX mutant partially restored particle production, consistent with a requirement for Gag-RNA binding in addition to Gag-Gag interactions. Together, these results support an RNA-binding requirement for Gag assembly, which relies on binding of RNA by MA or NC sequences to condense, organize, and stabilize the HIV-1 Gag-Gag interactions that form the virion.
Figures

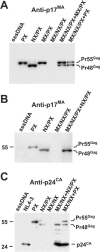
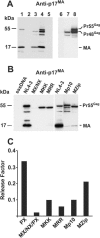
Similar articles
-
Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging.J Virol. 1998 Mar;72(3):1983-93. doi: 10.1128/JVI.72.3.1983-1993.1998. J Virol. 1998. PMID: 9499052 Free PMC article.
-
The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly.J Virol. 2009 Aug;83(15):7718-27. doi: 10.1128/JVI.00099-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457986 Free PMC article.
-
Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA.J Virol. 2000 Apr;74(7):3046-57. doi: 10.1128/jvi.74.7.3046-3057.2000. J Virol. 2000. PMID: 10708419 Free PMC article.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
-
How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain.Viruses. 2020 Aug 13;12(8):888. doi: 10.3390/v12080888. Viruses. 2020. PMID: 32823718 Free PMC article. Review.
Cited by
-
Differences and commonalities in plasma membrane recruitment of the two morphogenetically distinct retroviruses HIV-1 and MMTV.J Biol Chem. 2020 Jun 26;295(26):8819-8833. doi: 10.1074/jbc.RA119.011991. Epub 2020 May 8. J Biol Chem. 2020. PMID: 32385109 Free PMC article.
-
Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles.Nucleic Acids Res. 2011 Nov 1;39(20):8915-27. doi: 10.1093/nar/gkr577. Epub 2011 Jul 26. Nucleic Acids Res. 2011. PMID: 21791531 Free PMC article.
-
Retrovirus-specific differences in matrix and nucleocapsid protein-nucleic acid interactions: implications for genomic RNA packaging.J Virol. 2014 Jan;88(2):1271-80. doi: 10.1128/JVI.02151-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227839 Free PMC article.
-
Solution properties of murine leukemia virus gag protein: differences from HIV-1 gag.J Virol. 2011 Dec;85(23):12733-41. doi: 10.1128/JVI.05889-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917964 Free PMC article.
-
Characterization of a myristoylated, monomeric HIV Gag protein.Virology. 2009 May 10;387(2):341-52. doi: 10.1016/j.virol.2009.02.037. Epub 2009 Mar 12. Virology. 2009. PMID: 19285328 Free PMC article.
References
-
- Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

