Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies
- PMID: 16254605
- PMCID: PMC1270012
- DOI: 10.1371/journal.pgen.0010056
Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies
Abstract
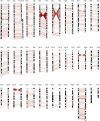
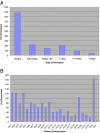
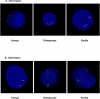
With a draft genome-sequence assembly for the chimpanzee available, it is now possible to perform genome-wide analyses to identify, at a submicroscopic level, structural rearrangements that have occurred between chimpanzees and humans. The goal of this study was to investigate chromosomal regions that are inverted between the chimpanzee and human genomes. Using the net alignments for the builds of the human and chimpanzee genome assemblies, we identified a total of 1,576 putative regions of inverted orientation, covering more than 154 mega-bases of DNA. The DNA segments are distributed throughout the genome and range from 23 base pairs to 62 mega-bases in length. For the 66 inversions more than 25 kilobases (kb) in length, 75% were flanked on one or both sides by (often unrelated) segmental duplications. Using PCR and fluorescence in situ hybridization we experimentally validated 23 of 27 (85%) semi-randomly chosen regions; the largest novel inversion confirmed was 4.3 mega-bases at human Chromosome 7p14. Gorilla was used as an out-group to assign ancestral status to the variants. All experimentally validated inversion regions were then assayed against a panel of human samples and three of the 23 (13%) regions were found to be polymorphic in the human genome. These polymorphic inversions include 730 kb (at 7p22), 13 kb (at 7q11), and 1 kb (at 16q24) fragments with a 5%, 30%, and 48% minor allele frequency, respectively. Our results suggest that inversions are an important source of variation in primate genome evolution. The finding of at least three novel inversion polymorphisms in humans indicates this type of structural variation may be a more common feature of our genome than previously realized.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
References
-
- Fujiyama A, Watanabe H, Toyoda A, Taylor TD, Itoh T, et al. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295:131–134. - PubMed
-
- Li WH, Saunders MA. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Hacia JG, Makalowski W, Edgemon K, Erdos MR, Robbins CM, et al. Evolutionary sequence comparisons using high-density oligonucleotide arrays. Nat Genet. 1998;18:155–158. - PubMed
-
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

