Causes of oncogenic chromosomal translocation
- PMID: 16257470
- PMCID: PMC1762911
- DOI: 10.1016/j.tig.2005.10.002
Causes of oncogenic chromosomal translocation
Abstract
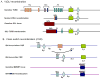
Non-random chromosomal translocations are frequently associated with a variety of cancers, particularly hematologic malignancies and childhood sarcomas. In addition to their diagnostic utility, chromosomal translocations are increasingly being used in the clinic to guide therapeutic decisions. However, the mechanisms that cause these translocations remain poorly understood. Illegitimate V(D)J recombination, class switch recombination, homologous recombination, non-homologous end-joining and genome fragile sites all have potential roles in the production of non-random chromosomal translocations. In addition, mutations in DNA-repair pathways have been implicated in the production of chromosomal translocations in humans, mice and yeast. Although initially surprising, the identification of these same oncogenic chromosomal translocations in peripheral blood from healthy individuals strongly suggests that the translocation is not sufficient to induce malignant transformation, and that complementary mutations are required to produce a frank malignancy.
Figures
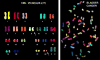

References
-
- Rabbitts TH. Chromosomal translocation master genes, mouse models and experimental therapeutics. Oncogene. 2001;20 (40):5763–5777. - PubMed
-
- Mitelman F, et al. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43 (4):350–366. - PubMed
-
- Chou WC, Dang CV. Acute promyelocytic leukemia: recent advances in therapy and molecular basis of response to arsenic therapies. Curr Opin Hematol. 2005;12 (1):1–6. - PubMed
-
- Hunger SP. Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis. Blood. 1996;87 (4):1211–1224. - PubMed
-
- Rubnitz JE, et al. 11q23 rearrangements in acute leukemia. Leukemia. 1996;10 (1):74–82. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

