Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans, an enzyme involved in lignin biosynthesis
- PMID: 16258008
- PMCID: PMC1283753
- DOI: 10.1104/pp.105.069674
Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans, an enzyme involved in lignin biosynthesis
Abstract
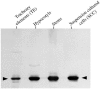
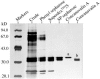
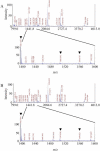
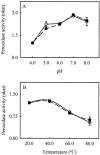
The major basic peroxidase from Zinnia elegans (ZePrx) suspension cell cultures was purified and cloned, and its properties and organ expression were characterized. The ZePrx was composed of two isoforms with a M(r) (determined by matrix-assisted laser-desorption ionization time of flight) of 34,700 (ZePrx34.70) and a M(r) of 33,440 (ZePrx33.44). Both isoforms showed absorption maxima at 403 (Soret band), 500, and 640 nm, suggesting that both are high-spin ferric secretory class III peroxidases. M(r) differences between them were due to the glycan moieties, and were confirmed from the total similarity of the N-terminal sequences (LSTTFYDTT) and by the 99.9% similarity of the tryptic fragment fingerprints obtained by reverse-phase nano-liquid chromatography. Four full-length cDNAs coding for these peroxidases were cloned. They only differ in the 5'-untranslated region. These differences probably indicate different ways in mRNA transport, stability, and regulation. According to the k(cat) and apparent K(m)(RH) values shown by both peroxidases for the three monolignols, sinapyl alcohol was the best substrate, the endwise polymerization of sinapyl alcohol by both ZePrxs yielding highly polymerized lignins with polymerization degrees > or =87. Western blots using anti-ZePrx34.70 IgGs showed that ZePrx33.44 was expressed in tracheary elements, roots, and hypocotyls, while ZePrx34.70 was only expressed in roots and young hypocotyls. None of the ZePrx isoforms was significantly expressed in either leaves or cotyledons. A neighbor-joining tree constructed for the four full-length cDNAs suggests that the four putative paralogous genes encoding the four cDNAs result from duplication of a previously duplicated ancestral gene, as may be deduced from the conserved nature and conserved position of the introns.
Figures





Similar articles
-
The promoter region of the Zinnia elegans basic peroxidase isoenzyme gene contains cis-elements responsive to nitric oxide and hydrogen peroxide.Planta. 2012 Aug;236(2):327-42. doi: 10.1007/s00425-012-1604-3. Epub 2012 Feb 24. Planta. 2012. PMID: 22362137
-
Post-translational modifications of the basic peroxidase isoenzyme from Zinnia elegans.Plant Mol Biol. 2007 Sep;65(1-2):43-61. doi: 10.1007/s11103-007-9197-0. Epub 2007 Jun 22. Plant Mol Biol. 2007. PMID: 17588152
-
Zinnia elegans uses the same peroxidase isoenzyme complement for cell wall lignification in both single-cell tracheary elements and xylem vessels.J Exp Bot. 2004 Feb;55(396):423-31. doi: 10.1093/jxb/erh036. J Exp Bot. 2004. PMID: 14739265
-
Characterization of the last step of lignin biosynthesis in Zinnia elegans suspension cell cultures.FEBS Lett. 2006 Aug 7;580(18):4311-6. doi: 10.1016/j.febslet.2006.06.088. Epub 2006 Jul 7. FEBS Lett. 2006. PMID: 16842784
-
From Zinnia to Arabidopsis: approaching the involvement of peroxidases in lignification.J Exp Bot. 2013 Sep;64(12):3499-518. doi: 10.1093/jxb/ert221. J Exp Bot. 2013. PMID: 23956408 Review.
Cited by
-
The promoter region of the Zinnia elegans basic peroxidase isoenzyme gene contains cis-elements responsive to nitric oxide and hydrogen peroxide.Planta. 2012 Aug;236(2):327-42. doi: 10.1007/s00425-012-1604-3. Epub 2012 Feb 24. Planta. 2012. PMID: 22362137
-
Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L).BMC Genomics. 2020 Jun 29;21(1):444. doi: 10.1186/s12864-020-06828-z. BMC Genomics. 2020. PMID: 32600251 Free PMC article.
-
Characterization of Peroxidase and Laccase Gene Families and In Silico Identification of Potential Genes Involved in Upstream Steps of Lignan Formation in Sesame.Life (Basel). 2022 Aug 8;12(8):1200. doi: 10.3390/life12081200. Life (Basel). 2022. PMID: 36013379 Free PMC article.
-
Transcriptional Roadmap to Seasonal Variation in Wood Formation of Norway Spruce.Plant Physiol. 2018 Apr;176(4):2851-2870. doi: 10.1104/pp.17.01590. Epub 2018 Feb 27. Plant Physiol. 2018. PMID: 29487121 Free PMC article.
-
Regulation of Lignin Biosynthesis by Post-translational Protein Modifications.Front Plant Sci. 2020 Jul 2;11:914. doi: 10.3389/fpls.2020.00914. eCollection 2020. Front Plant Sci. 2020. PMID: 32714349 Free PMC article. Review.
References
-
- Anterola AM, Jeon J, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda. J Biol Chem 227: 18272–18280 - PubMed
-
- Anterola AM, Van Rensburg H, Van Hereden PS, Davin LB, Lewis NG (1999) Multi-site modulation of flux during monolignol formation in loblolly pine (Pinus taeda). Biochem Biophys Res Commun 261: 652–657 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

