Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis
- PMID: 16258155
- PMCID: PMC1867550
- DOI: 10.1016/S1525-1578(10)60590-9
Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis
Abstract
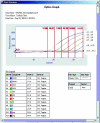
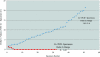
Speed is of the essence when evaluating an infant with symptoms consistent with sepsis. Because of the high morbidity and mortality associated with neonatal sepsis, infants receive multiple, broad-spectrum antibiotics before receiving finalized blood culture results. Incorporating an additional, reliable, yet rapid assay to detect bacteria directly from blood would facilitate timely diagnosis and appropriate care. To this end, we designed a real-time polymerase chain reaction (PCR) assay targeting the highly conserved 380 bases of 16S rDNA. DNA was extracted from whole-blood samples using a Qiagen column. The limit of detection for the TaqMan-based assay, using a Smartcycler instrument, was 40, 50, or 2000 colony-forming units per milliliter of blood (CFU/ml) of Escherichia coli, group B Streptococcus, and Listeria monocytogenes, respectively, when white blood cell counts were below 39,000/microl. Implementing this approach requires less than 4 hours for both sample preparation and real-time PCR compared with 1 to 2 days to detect growth in culture or 5 days to finalize no-growth culture results. There was an overall agreement between the results of culture and real-time PCR of 94.1% (80 of 85) in this study. These results suggest that molecular techniques can augment culture-based methods for diagnosing neonatal sepsis, especially in infants whose mothers have received intrapartum antibiotic prophylaxis.
Figures
References
-
- Cerase PA. Neonatal sepsis. J Perinat Neonat Nurs. 1989;3:48–57. - PubMed
-
- Gerdes JS. Clinicopathologic approach to the diagnosis of neonatal sepsis. Clin Perinatol. 1991;18:361–381. - PubMed
-
- Klein JO. Philadelphia: W.B. Saunders,; Bacterial sepsis and meningitis. Infectious Diseases of the Fetus and Newborn. 2001:943–998.
-
- Witek-Janusek L, Cusack C. Neonatal sepsis: confronting the challenge. Crit Care Nurs Clin North Am. 1994;6:405–419. - PubMed
-
- Chadwick EG, Yogev R, Shulman ST. Combination antibiotic therapy in pediatrics. Am J Med. 1986;80:166–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

