Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome
- PMID: 16260473
- PMCID: PMC1275586
- DOI: 10.1093/nar/gki925
Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome
Abstract
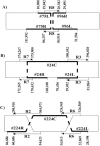
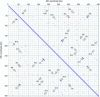
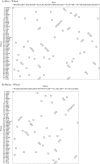
The application of a new gene-based strategy for sequencing the wheat mitochondrial genome shows its structure to be a 452 528 bp circular molecule, and provides nucleotide-level evidence of intra-molecular recombination. Single, reciprocal and double recombinant products, and the nucleotide sequences of the repeats that mediate their formation have been identified. The genome has 55 genes with exons, including 35 protein-coding, 3 rRNA and 17 tRNA genes. Nucleotide sequences of seven wheat genes have been determined here for the first time. Nine genes have an exon-intron structure. Gene amplification responsible for the production of multicopy mitochondrial genes, in general, is species-specific, suggesting the recent origin of these genes. About 16, 17, 15, 3.0 and 0.2% of wheat mitochondrial DNA (mtDNA) may be of genic (including introns), open reading frame, repetitive sequence, chloroplast and retro-element origin, respectively. The gene order of the wheat mitochondrial gene map shows little synteny to the rice and maize maps, indicative that thorough gene shuffling occurred during speciation. Almost all unique mtDNA sequences of wheat, as compared with rice and maize mtDNAs, are redundant DNA. Features of the gene-based strategy are discussed, and a mechanistic model of mitochondrial gene amplification is proposed.
Figures




References
-
- Miller R.J., Koeppe D.E. Southern corn leaf blight: susceptible and resistant mitochondria. Science. 1971;173:67–69. - PubMed
-
- Cooper G.M. The Cell—A Molecular Approach, 2nd edn. Sunderland: Sinauer Assoc., Inc.; 2000.
-
- Oda K., Yamato K., Ohta E., Nakamura Y., Takemura M., Nozato N., Akashi K., Kanegae T., Ogura Y., Kohchi T., et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA, a primitive form of plant mitochondrial genome. J. Mol. Biol. 1992;223:1–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

