Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae
- PMID: 16260619
- PMCID: PMC1280264
- DOI: 10.1128/MCB.25.22.10060-10070.2005
Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae
Abstract
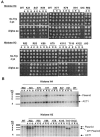
The biological significance of recently described modifiable residues in the globular core of the bovine nucleosome remains elusive. We have mapped these modification sites onto the Saccharomyces cerevisiae histones and used a genetic approach to probe their potential roles both in heterochromatic regions of the genome and in the DNA repair response. By mutating these residues to mimic their modified and unmodified states, we have generated a total of 39 alleles affecting 14 residues in histones H3 and H4. Remarkably, despite the apparent evolutionary pressure to conserve these near-invariant histone amino acid sequences, the vast majority of mutant alleles are viable. However, a subset of these variant proteins elicit an effect on transcriptional silencing both at the ribosomal DNA locus and at telomeres, suggesting that posttranslational modification(s) at these sites regulates formation and/or maintenance of heterochromatin. Furthermore, we provide direct mass spectrometry evidence for the existence of histone H3 K56 acetylation in yeast. We also show that substitutions at histone H4 K91, K59, S47, and R92 and histone H3 K56 and K115 lead to hypersensitivity to DNA-damaging agents, linking the significance of the chemical identity of these modifiable residues to DNA metabolism. Finally, we allude to the possible molecular mechanisms underlying the effects of these modifications.
Figures





References
-
- Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
-
- Chang, C. H., and D. S. Luse. 1997. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 272:23427-23434. - PubMed
-
- Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656-660. - PubMed
-
- Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. - PubMed
-
- Cost, G. J., and J. D. Boeke. 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12:939-941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
