A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis
- PMID: 16260724
- PMCID: PMC1283476
- DOI: 10.1073/pnas.0508448102
A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis
Erratum in
- Proc Natl Acad Sci U S A. 2006 Jan 17;103(3):825
Abstract
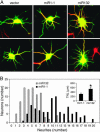
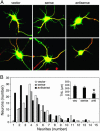
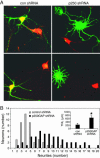
MicroRNAs (miRNAs) regulate cellular fate by controlling the stability or translation of mRNA transcripts. Although the spatial and temporal patterning of miRNA expression is tightly controlled, little is known about signals that induce their expression nor mechanisms of their transcriptional regulation. Furthermore, few miRNA targets have been validated experimentally. The miRNA, miR132, was identified through a genome-wide screen as a target of the transcription factor, cAMP-response element binding protein (CREB). miR132 is enriched in neurons and, like many neuronal CREB targets, is highly induced by neurotrophins. Expression of miR132 in cortical neurons induced neurite outgrowth. Conversely, inhibition of miR132 function attenuated neuronal outgrowth. We provide evidence that miR132 regulates neuronal morphogenesis by decreasing levels of the GTPase-activating protein, p250GAP. These data reveal that a CREB-regulated miRNA regulates neuronal morphogenesis by responding to extrinsic trophic cues.
Figures




References
-
- Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605-623. - PubMed
-
- Gonzalez, G. A. & Montminy, M. R. (1989) Cell 59, 675-680. - PubMed
-
- Chrivia, J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365, 855-859. - PubMed
-
- Vo, N. & Goodman, R. H. (2001) J. Biol. Chem. 276, 13505-13508. - PubMed
-
- Conkright, M. D., Canettieri, G., Screaton, R., Guzman, E., Miraglia, L., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 12, 413-423. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

