An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites
- PMID: 16260729
- PMCID: PMC1283470
- DOI: 10.1073/pnas.0508335102
An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites
Abstract
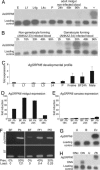
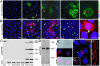
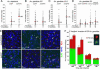
We have functionally analyzed the orthologous SRPN6 genes from Anopheles stephensi and Anopheles gambiae using phylogenetic, molecular, reverse genetic, and cell biological tools. The results strongly implicate SRPN6 in the innate immune response against Plasmodium. This gene belongs to a mosquito-specific gene cluster including three additional Anopheles serpins. SRPN6 expression is induced by Escherichia coli and both rodent and human malaria parasites. The gene is specifically expressed in midgut cells invaded by Plasmodium ookinetes and in circulating and attached hemocytes. Knockdown of SRPN6 expression by RNA interference in susceptible An. stephensi leads to substantially increased parasite numbers, whereas depletion in susceptible An. gambiae delays progression of parasite lysis without affecting the number of developing parasites. However, the An. gambiae SRPN6 knockdown increases the number of melanized parasites in the L3-5 refractory strain and in susceptible G3 mosquitoes depleted of CTL4. These results indicate that AsSRPN6 is involved in the parasite-killing process, whereas AgSRPN6 acts on parasite clearance by inhibiting melanization and/or promoting parasite lysis. We propose that these observed phenotypic differences are due to changed roles of the respective target serine proteases in the two mosquito species.
Figures

References
-
- Niare, O., Markianos, K., Volz, J., Oduol, F., Toure, A., Bagayoko, M., Sangare, D., Traore, S. F., Wang, R., Blass, C., et al. (2002) Science 298, 213-216. - PubMed
-
- Sinden, R. E. (2002) Cell. Microbiol. 4, 713-724. - PubMed
-
- Collins, F. H., Sakai, R. K., Vernick, K. D., Paskewitz, S., Seeley, D. C., Miller, L. H., Collins, W. E., Campbell, C. C. & Gwadz, R. W. (1986) Science 234, 607-610. - PubMed
-
- Vernick, K. D., Fujioka, H., Seeley, D. C., Tandler, B., Aikawa, M. & Miller, L. H. (1995) Exp. Parasitol. 80, 583-595. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

