Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
- PMID: 16260736
- PMCID: PMC1297672
- DOI: 10.1073/pnas.0506924102
Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Abstract
Integrase (IN) is an essential retroviral enzyme, and human transcriptional coactivator p75, which is also referred to as lens epithelium-derived growth factor (LEDGF), is the dominant cellular binding partner of HIV-1 IN. Here, we report the crystal structure of the dimeric catalytic core domain of HIV-1 IN complexed to the IN-binding domain of LEDGF. Previously identified LEDGF hotspot residues anchor the protein to both monomers at the IN dimer interface. The principal structural features of IN that are recognized by the host factor are the backbone conformation of residues 168-171 from one monomer and a hydrophobic patch that is primarily comprised of alpha-helices 1 and 3 of the second IN monomer. Inspection of diverse retroviral primary and secondary sequence elements helps to explain the apparent lentiviral tropism of the LEDGF-IN interaction. Because the lethal phenotypes of HIV-1 mutant viruses unable to interact with LEDGF indicate that IN function is highly sensitive to perturbations of the structure around the LEDGF-binding site, we propose that small molecule inhibitors of the protein-protein interaction might similarly disrupt HIV-1 replication.
Figures

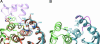
Comment in
-
Seeing is believing: structure of the catalytic domain of HIV-1 integrase in complex with human LEDGF/p75.Proc Natl Acad Sci U S A. 2005 Dec 6;102(49):17543-4. doi: 10.1073/pnas.0509078102. Epub 2005 Nov 28. Proc Natl Acad Sci U S A. 2005. PMID: 16314581 Free PMC article. No abstract available.
References
-
- Craigie, R. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 613–630.
-
- Turlure, F., Devroe, E., Silver, P. A. & Engelman, A. (2004) Front. Biosci. 9, 3187–3208. - PubMed
-
- Bushman, F. D. (2003) Cell 115, 135–138. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

