Disruption of an intermonomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils
- PMID: 16260757
- PMCID: PMC2253254
- DOI: 10.1110/ps.051622005
Disruption of an intermonomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils
Abstract
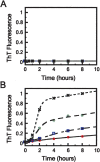
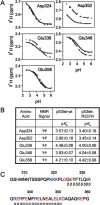
We describe in molecular detail how disruption of an intermonomer salt bridge (Arg337-Asp352) leads to partial destabilization of the p53 tetramerization domain and a dramatically increased propensity to form amyloid fibrils. At pH 4.0 and 37 degrees C, a p53 tetramerization domain mutant (p53tet-R337H), associated with adrenocortical carcinoma in children, readily formed amyloid fibrils, while the wild-type (p53tet-wt) did not. We characterized these proteins by equilibrium denaturation, 13C(alpha) secondary chemical shifts, (1H)-15N heteronuclear NOEs, and H/D exchange. Although p53tet-R337H was thermodynamically less stable, NMR data indicated that the two proteins had similar secondary structure and molecular dynamics. NMR derived pK(a) values indicated that at low pH the R337H mutation partially disrupted an intermonomer salt bridge. Backbone H/D exchange results showed that for at least a small population of p53tet-R337H molecules disruption of this salt bridge resulted in partial destabilization of the protein. It is proposed that this decrease in p53tet-R337H stability resulted in an increased propensity to form amyloid fibrils.
Figures



References
-
- Booth, D.R., Sunde, M., Bellotti, V., Robinson, C.V., Hutchinson, W.L., Fraser, P.E., Hawkins, P.N., Dobson, C.M., Radford, S.E., Blake, C.C., et al. 1997. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature 385 787–793. - PubMed
-
- Bosshard, H.R., Marti, D.N., and Jelesarov, I. 2004. Protein stabilization by salt bridges: Concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recognit. 17 1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

