The family feud: turning off Sp1 by Sp1-like KLF proteins
- PMID: 16266294
- PMCID: PMC1317658
- DOI: 10.1042/BJ20051234
The family feud: turning off Sp1 by Sp1-like KLF proteins
Abstract
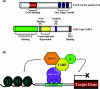
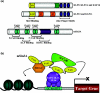
Sp1 is one of the best characterized transcriptional activators. The biological importance of Sp1 is underscored by the fact that several hundreds of genes are thought to be regulated by this protein. However, during the last 5 years, a more extended family of Sp1-like transcription factors has been identified and characterized by the presence of a conserved DNA-binding domain comprising three Krüppel-like zinc fingers. Each distinct family member differs in its ability to regulate transcription, and, as a consequence, to influence cellular processes. Specific activation and repression domains located within the N-terminal regions of these proteins are responsible for these differences by facilitating interactions with various co-activators and co-repressors. The present review primarily focuses on discussing the structural, biochemical and biological functions of the repressor members of this family of transcription factors. The existence of these transcriptional repressors provides a tightly regulated mechanism for silencing a large number of genes that are already known to be activated by Sp1.
Figures

References
-
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell (Cambridge, Mass.) 1983;35:79–87. - PubMed
-
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell (Cambridge, Mass.) 1987;51:1079–1090. - PubMed
-
- Gidoni D., Kadonaga J. T., Barrera-Saldana H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science (Washington, D.C.) 1985;230:511–517. - PubMed
-
- Giglioni B., Comi P., Ronchi A., Mantovani R., Ottolenghi S. The same nuclear proteins bind the proximal CACCC box of the human β-globin promoter and a similar sequence in the enhancer. Biochem. Biophys. Res. Commun. 1989;164:149–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

