Gene expression patterns and catalytic properties of UDP-D-glucose 4-epimerases from barley (Hordeum vulgare L.)
- PMID: 16266295
- PMCID: PMC1386009
- DOI: 10.1042/BJ20051329
Gene expression patterns and catalytic properties of UDP-D-glucose 4-epimerases from barley (Hordeum vulgare L.)
Abstract
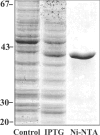
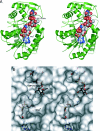
UGE (UDP-Glc 4-epimerase or UDP-Gal 4-epimerase; EC 5.1.3.2) catalyses the interconversion of UDP-Gal and UDP-Glc. Both nucleotide sugars act as activated sugar donors for the biosynthesis of cell wall polysaccharides such as cellulose, xyloglucans, (1,3;1,4)-beta-D-glucan and pectins, together with other biologically significant compounds including glycoproteins and glycolipids. Three members of the HvUGE (barley UGE) gene family, designated HvUGE1, HvUGE2 and HvUGE3, have been characterized. Q-PCR (quantitative real-time PCR) showed that HvUGE1 mRNA was most abundant in leaf tips and mature roots, but its expression levels were relatively low in basal leaves and root tips. The HvUGE2 gene was transcribed at significant levels in all organs examined, while HvUGE3 mRNA levels were very low in all the organs. Heterologous expression of a near full-length cDNA confirmed that HvUGE1 encodes a functional UGE. A non-covalently bound NAD+ was released from the enzyme after denaturing with aqueous ethanol and was identified by its spectrophotometric properties and by electrospray ionization MS. The K(m) values were 40 microM for UDP-Gal and 55 muM for UDP-Glc. HvUGE also catalyses the interconversion of UDP-GalNAc and UDP-GlcNAc, although it is not known if this has any biological significance. A three-dimensional model of the HvUGE revealed that its overall structural fold is highly conserved compared with the human UGE and provides a structural rationale for its ability to bind UDP-GlcNAc.
Figures

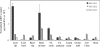

References
-
- Reiter W. D., Vanzin G. F. Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 2001;47:95–113. - PubMed
-
- Seifert G. J. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr. Opin. Plant Biol. 2004;7:277–284. - PubMed
-
- Bauer A. J., Rayment I., Frey P. A., Holden H. M. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 Å resolution. Proteins. 1992;12:372–381. - PubMed
-
- Barat B., Bhattacharyya D. UDP-galactose 4-epimerase from Escherichia coli: formation of catalytic site during reversible folding. Arch. Biochem. Biophys. 2001;391:188–196. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

