The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans
- PMID: 16267276
- PMCID: PMC1345667
- DOI: 10.1091/mbc.e05-06-0502
The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans
Abstract
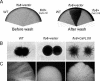
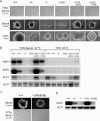
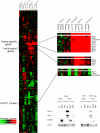
The transcription factor Flo8 is essential for filamentous growth in Saccharomyces cerevisiae and is regulated under the cAMP/protein kinase A (PKA) pathway. To determine whether a similar pathway/regulation exists in Candida albicans, we have cloned C. albicans FLO8 by its ability to complement S. cerevisiae flo8. Deleting FLO8 in C. albicans blocked hyphal development and hypha-specific gene expression. The flo8/flo8 mutant is avirulent in a mouse model of systemic infection. Genome-wide transcription profiling of efg1/efg1 and flo8/flo8 using a C. albicans DNA microarray suggests that Flo8 controls subsets of Efg1-regulated genes. Most of these genes are hypha specific, including HGC1 and IHD1. We also show that Flo8 interacts with Efg1 in yeast and hyphal cells by in vivo immunoprecipitation. Similar to efg1/efg1, flo8/flo8 and cdc35/cdc35 show enhanced hyphal growth under an embedded growth condition. Our results suggest that Flo8 may function downstream of the cAMP/PKA pathway, and together with Efg1, regulates the expression of hypha-specific genes and genes that are important for the virulence of C. albicans.
Figures





References
-
- Bockmuhl, D. P., Krishnamurthy, S., Gerads, M., Sonneborn, A., and Ernst, J. F. (2001). Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42, 1243–1257. - PubMed
-
- Brown, D. H., Jr., Giusani, A. D., Chen, X., and Kumamoto, C. A. (1999). Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34, 651–662. - PubMed
-
- Cassola, A., Parrot, M., Silberstein, S., Magee, B. B., Passeron, S., Giasson, L., and Cantore, M. L. (2004). Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3, 190–199. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

