Genetic composition of the Bacillus subtilis SOS system
- PMID: 16267290
- PMCID: PMC1280312
- DOI: 10.1128/JB.187.22.7655-7666.2005
Genetic composition of the Bacillus subtilis SOS system
Abstract
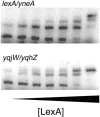
The SOS response in bacteria includes a global transcriptional response to DNA damage. DNA damage is sensed by the highly conserved recombination protein RecA, which facilitates inactivation of the transcriptional repressor LexA. Inactivation of LexA causes induction (derepression) of genes of the LexA regulon, many of which are involved in DNA repair and survival after DNA damage. To identify potential RecA-LexA-regulated genes in Bacillus subtilis, we searched the genome for putative LexA binding sites within 300 bp upstream of the start codons of all annotated open reading frames. We found 62 genes that could be regulated by putative LexA binding sites. Using mobility shift assays, we found that LexA binds specifically to DNA in the regulatory regions of 54 of these genes, which are organized in 34 putative operons. Using DNA microarray analyses, we found that 33 of the genes with LexA binding sites exhibit RecA-dependent induction by both mitomycin C and UV radiation. Among these 33 SOS genes, there are 22 distinct LexA binding sites preceding 18 putative operons. Alignment of the distinct LexA binding sites reveals an expanded consensus sequence for the B. subtilis operator: 5'-CGAACATATGTTCG-3'. Although the number of genes controlled by RecA and LexA in B. subtilis is similar to that of Escherichia coli, only eight B. subtilis RecA-dependent SOS genes have homologous counterparts in E. coli.
Figures
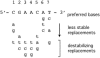



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

