maxdLoad2 and maxdBrowse: standards-compliant tools for microarray experimental annotation, data management and dissemination
- PMID: 16269077
- PMCID: PMC1298287
- DOI: 10.1186/1471-2105-6-264
maxdLoad2 and maxdBrowse: standards-compliant tools for microarray experimental annotation, data management and dissemination
Abstract
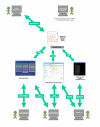
Background: maxdLoad2 is a relational database schema and Java application for microarray experimental annotation and storage. It is compliant with all standards for microarray meta-data capture; including the specification of what data should be recorded, extensive use of standard ontologies and support for data exchange formats. The output from maxdLoad2 is of a form acceptable for submission to the ArrayExpress microarray repository at the European Bioinformatics Institute. maxdBrowse is a PHP web-application that makes contents of maxdLoad2 databases accessible via web-browser, the command-line and web-service environments. It thus acts as both a dissemination and data-mining tool.
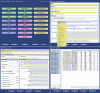
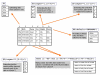
Results: maxdLoad2 presents an easy-to-use interface to an underlying relational database and provides a full complement of facilities for browsing, searching and editing. There is a tree-based visualization of data connectivity and the ability to explore the links between any pair of data elements, irrespective of how many intermediate links lie between them. Its principle novel features are: the flexibility of the meta-data that can be captured, the tools provided for importing data from spreadsheets and other tabular representations, the tools provided for the automatic creation of structured documents, the ability to browse and access the data via web and web-services interfaces. Within maxdLoad2 it is very straightforward to customise the meta-data that is being captured or change the definitions of the meta-data. These meta-data definitions are stored within the database itself allowing client software to connect properly to a modified database without having to be specially configured. The meta-data definitions (configuration file) can also be centralized allowing changes made in response to revisions of standards or terminologies to be propagated to clients without user intervention.maxdBrowse is hosted on a web-server and presents multiple interfaces to the contents of maxd databases. maxdBrowse emulates many of the browse and search features available in the maxdLoad2 application via a web-browser. This allows users who are not familiar with maxdLoad2 to browse and export microarray data from the database for their own analysis. The same browse and search features are also available via command-line and SOAP server interfaces. This both enables scripting of data export for use embedded in data repositories and analysis environments, and allows access to the maxd databases via web-service architectures.
Conclusion: maxdLoad2 http://www.bioinf.man.ac.uk/microarray/maxd/ and maxdBrowse http://dbk.ch.umist.ac.uk/maxdBrowse are portable and compatible with all common operating systems and major database servers. They provide a powerful, flexible package for annotation of microarray experiments and a convenient dissemination environment. They are available for download and open sourced under the Artistic License.
Figures




Similar articles
-
MiMiR--an integrated platform for microarray data sharing, mining and analysis.BMC Bioinformatics. 2008 Sep 18;9:379. doi: 10.1186/1471-2105-9-379. BMC Bioinformatics. 2008. PMID: 18801157 Free PMC article.
-
Performing statistical analyses on quantitative data in Taverna workflows: an example using R and maxdBrowse to identify differentially-expressed genes from microarray data.BMC Bioinformatics. 2008 Aug 7;9:334. doi: 10.1186/1471-2105-9-334. BMC Bioinformatics. 2008. PMID: 18687127 Free PMC article.
-
Gene Expression Browser: large-scale and cross-experiment microarray data integration, management, search & visualization.BMC Bioinformatics. 2010 Aug 20;11:433. doi: 10.1186/1471-2105-11-433. BMC Bioinformatics. 2010. PMID: 20727159 Free PMC article.
-
Storage and retrieval of microarray data and open source microarray database software.Mol Biotechnol. 2005 Jul;30(3):239-51. doi: 10.1385/MB:30:3:239. Mol Biotechnol. 2005. PMID: 15988049 Review.
-
A guide for integration of proteomic data standards into laboratory workflows.Proteomics. 2013 Feb;13(3-4):480-92. doi: 10.1002/pmic.201200268. Epub 2013 Jan 15. Proteomics. 2013. PMID: 23319203 Review.
Cited by
-
Towards large-scale sample annotation in gene expression repositories.BMC Bioinformatics. 2009 Sep 17;10 Suppl 9(Suppl 9):S9. doi: 10.1186/1471-2105-10-S9-S9. BMC Bioinformatics. 2009. PMID: 19761579 Free PMC article.
-
MeMo: a hybrid SQL/XML approach to metabolomic data management for functional genomics.BMC Bioinformatics. 2006 Jun 5;7:281. doi: 10.1186/1471-2105-7-281. BMC Bioinformatics. 2006. PMID: 16753052 Free PMC article.
-
Gene expression profiling in a mouse model for African trypanosomiasis.Genes Immun. 2006 Dec;7(8):667-79. doi: 10.1038/sj.gene.6364345. Epub 2006 Oct 26. Genes Immun. 2006. PMID: 17066074 Free PMC article.
-
A reproducible approach to high-throughput biological data acquisition and integration.PeerJ. 2015 Mar 31;3:e791. doi: 10.7717/peerj.791. eCollection 2015. PeerJ. 2015. PMID: 26157642 Free PMC article.
-
Claudin 13, a member of the claudin family regulated in mouse stress induced erythropoiesis.PLoS One. 2010 Sep 10;5(9):e12667. doi: 10.1371/journal.pone.0012667. PLoS One. 2010. PMID: 20844758 Free PMC article.
References
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. - DOI - PubMed
-
- Group OM. Gene Expression Specification v1.1. The Object Management Group, Inc; http://www.omg.org/cgi-bin/doc?formal/03-10-01 3 AD.
-
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks WL, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert CJJ, Brazma A. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biology. 2002;3:RESEARCH0046. doi: 10.1186/gb-2002-3-9-research0046. - DOI - PMC - PubMed
-
- MADAM. 2005. http://www.tm4.org/madam.html
MeSH terms
LinkOut - more resources
Full Text Sources

