Identifying components of the NF-kappaB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis
- PMID: 16269728
- PMCID: PMC1287678
- DOI: 10.1128/AEM.71.11.6934-6946.2005
Identifying components of the NF-kappaB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis
Abstract
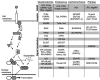
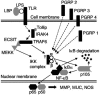
The Toll/NF-kappaB pathway is a common, evolutionarily conserved innate immune pathway that modulates the responses of animal cells to microbe-associated molecular patterns (MAMPs). Because MAMPs have been implicated as critical elements in the signaling of symbiont-induced development, an expressed sequence tag library from the juvenile light organ of Euprymna scolopes was used to identify members of the Toll/NF-kappaB pathway. Full-length transcripts were identified by using 5' and 3' RACE PCR. Seven transcripts critical for MAMP-induced triggering of the Toll/NF-kappaB phosphorylation cascade have been identified, including receptors, signal transducers, and a transcription factor. Further investigations should elucidate the role of the Toll/NF-kappaB pathway in the initiation of the beneficial symbiosis between E. scolopes and Vibrio fischeri.
Figures







References
-
- Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. - PubMed
-
- Aguinaldo, A. M., J. M. Turbeville, L. S. Linford, M. C. Rivera, J. R. Garey, R. A. Raff, and J. A. Lake. 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387:489-493. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anton, P. M., V. Theodorou, S. Roy, J. Fioramonti, and L. Bueno. 2002. Pathways involved in mild gastrointestinal inflammation induced by a low level exposure to a food contaminant. Dig. Dis. Sci. 47:1308-1315. - PubMed
-
- Asai, T., G. Tena, J. Plotnikova, M. R. Willmann, W. L. Chiu, L. Gomez-Gomez, T. Boller, F. M. Ausubel, and J. Sheen. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977-983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

