Community composition of a hypersaline endoevaporitic microbial mat
- PMID: 16269778
- PMCID: PMC1287706
- DOI: 10.1128/AEM.71.11.7352-7365.2005
Community composition of a hypersaline endoevaporitic microbial mat
Abstract
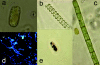
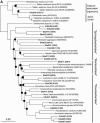
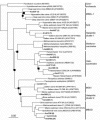
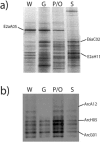
A hypersaline, endoevaporitic microbial community in Eilat, Israel, was studied by microscopy and by PCR amplification of genes for 16S rRNA from different layers. In terms of biomass, the oxygenic layers of the community were dominated by Cyanobacteria of the Halothece, Spirulina, and Phormidium types, but cell counts (based on 4',6'-diamidino-2-phenylindole staining) and molecular surveys (clone libraries of PCR-amplified genes for 16S rRNA) showed that oxygenic phototrophs were outnumbered by the other constituents of the community, including chemotrophs and anoxygenic phototrophs. Bacterial clone libraries were dominated by phylotypes affiliated with the Bacteroidetes group and both photo- and chemotrophic groups of alpha-proteobacteria. Green filaments related to the Chloroflexi were less abundant than reported from hypersaline microbial mats growing at lower salinities and were only detected in the deepest part of the anoxygenic phototrophic zone. Also detected were nonphototrophic gamma- and delta-proteobacteria, Planctomycetes, the TM6 group, Firmicutes, and Spirochetes. Several of the phylotypes showed a distinct vertical distribution in the crust, suggesting specific adaptations to the presence or absence of oxygen and light. Archaea were less abundant than Bacteria, their diversity was lower, and the community was less stratified. Detected archaeal groups included organisms affiliated with the Methanosarcinales, the Halobacteriales, and uncultured groups of Euryarchaeota.
Figures



References
-
- Benlloch, S., S. G. Acinas, J. Antón, A. López-López, S. P. Luz, and F. Rodríguez-Valera. 2001. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb. Ecol. 41:12-19. - PubMed
-
- Benlloch, S., A. López-López, E. O. Casamayor, L. Øvreås, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Joint, F. Thingstad, C. Pedrós-Alió, and F. Rodríguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. - PubMed
-
- Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ. Microbiol. 2:227-237. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

