Characterization of human NSCLC cell line with innate etoposide-resistance mediated by cytoplasmic localization of topoisomerase II alpha
- PMID: 16271071
- PMCID: PMC11158927
- DOI: 10.1111/j.1349-7006.2005.00111.x
Characterization of human NSCLC cell line with innate etoposide-resistance mediated by cytoplasmic localization of topoisomerase II alpha
Abstract
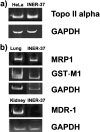
Topoisomerase (topo) II alpha is a target for many chemotherapeutic agents in clinical use. In tumor cells resistant to topo II poisons, there have been descriptions of quantitative and qualitative alterations involved in this enzyme. More recently, the cytoplasmic localization of topo II alpha has been described as a mechanism to confer drug resistance. Here, we report the characterization of a human non-small-cell lung cancer cell line, INER-37, which shows an innate resistance to etoposide. In this cell line, etoposide resistance was directly associated with the expression of topo II alpha resident mainly in the cytoplasmic region. At the molecular level, INER-37 cells carry on a heterozygous gene deletion, transcribing two different topo II alpha mRNAs: 4.8 kb and 2.0 kb. The bigger 4.8 kb mRNA (missing 1.3 kb of 3' mRNA and including the untranslated region) is translated into a truncated cytoplasmic protein of approximately 160 kDa. The protein truncation affects at least 96 amino acids in the COOH-terminal region where the more proximal bipartite nuclear localization signal is located. The INER-37 cell line is the first cancer cell line reported with an innate mutation affecting the 3'-end region of the topo II alpha gene that confers a cytoplasmic localization of the enzyme and therefore an increased resistance to etoposide.
Figures




References
-
- Wang JC. DNA topoisomerases. Annu Rev Biochem 1996; 65: 635–92. - PubMed
-
- Champoux JJ. DNA topoisomerases: Structure, function and mechanism. Annu Rev Biochem 2001; 70: 369–413. - PubMed
-
- Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 1989; 58: 350–75. - PubMed
-
- Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta 1998; 1400: 139–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

