Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis
- PMID: 16272474
- PMCID: PMC1287802
- DOI: 10.1128/JCM.43.11.5483-5490.2005
Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis
Abstract
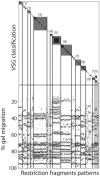
Pulsed-field gel electrophoresis (PFGE) has been the typing method of choice for strain identification in epidemiological studies of several bacterial species of medical importance. The usual procedure for the comparison of strains and assignment of strain type and subtype relies on visual assessment of band difference number, followed by an incremental assignment to the group hosting the most similar type previously seen. Band-based similarity coefficients, such as the Dice or the Jaccard coefficient, are then used for dendrogram construction, which provides a quantitative assessment of strain similarity. PFGE type assignment is based on the definition of a threshold linkage value, below which strains are assigned to the same group. This is typically performed empirically by inspecting the hierarchical cluster analysis dendrogram containing the strains of interest. This approach has the problem that the threshold value selected is dependent on the linkage method used for dendrogram construction. Furthermore, the use of a linkage method skews the original similarity values between strains. In this paper we assess the goodness of classification of several band-based similarity coefficients by comparing it with the band difference number for PFGE type and subtype classification using receiver operating characteristic curves. The procedure described was applied to a collection of PFGE results for 1,798 isolates of Streptococcus pneumoniae, which documented 96 types and 396 subtypes. The band-based similarity coefficients were found to perform equally well for type classification, but with different proportions of false-positive and false-negative classifications in their minimal false discovery rate when they were used for subtype classification.
Figures



References
-
- Baldi, P., S. Brunak, Y. Chauvin, C. A. Andersen, and H. Nielsen. 2000. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 16:412-424. - PubMed
-
- Bradley, A. P. 1997. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition 30:1145-1159.
-
- Bronzwaer, S. L., U. Buchholz, J. L. Kool, J. Monen, and P. Schrijnemakers. 2001. EARSS activities and results: update. Euro. Surveill. 6:2-5. - PubMed
-
- Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

