Analytical verification of a PCR assay for identification of Bordetella avium
- PMID: 16272488
- PMCID: PMC1287781
- DOI: 10.1128/JCM.43.11.5567-5573.2005
Analytical verification of a PCR assay for identification of Bordetella avium
Abstract
Bordetella avium is the etiologic agent of turkey coryza or bordetellosis, a respiratory disease responsible for substantial economic losses to the turkey industry. At present, identification of this bacterium relies on isolation and biochemical testing. Although a PCR for the detection of B. avium was proposed a number of years ago, lack of analytical verification precludes its use as a diagnostic tool. Furthermore, a number of details pertaining to the reaction conditions used are missing or unclear. In the present study we have identified an optimal set of PCR conditions for use with the previously described primer pair and determined the limit of detection under these conditions to be approximately 20 pg. Assay sensitivity is 100%, based on an analysis of 72 B. avium isolates from diverse geographic locations and covering a time span of at least 25 years. Evaluation of a separate group of 87 bacterial isolates from poultry, comprising both gram-positive and gram-negative commensals and pathogens representing 11 genera, demonstrated an assay specificity of 98.8%. Reproducibility is 100% using either purified genomic DNA or boiled cell lysates less than 3 days old. Sequence analysis of the B. avium PCR amplicons identified only three occasional sequence polymorphisms. These data indicate the B. avium PCR assay can provide clinically significant results.
Figures


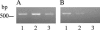
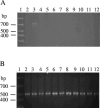

References
-
- Blackall, P. J., and C. M. Doheny. 1987. Isolation and characterisation of Bordetella avium and related species and an evaluation of their role in respiratory disease in poultry. Aust. Vet. J. 64:235-239. - PubMed
-
- Blackall, P. J., and J. G. Farrah. 1986. An evaluation of two methods of substrate alkalinization for the identification of Bordetella avium and other similar organisms. Vet. Microbiol. 11:301-306. - PubMed
-
- Cai, H., M. Archambault, and J. F. Prescott. 2003. 16S ribosomal RNA sequence-based identification of veterinary clinical bacteria. J. Vet. Diagn. Investig. 15:465-469. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

