Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge
- PMID: 16272491
- PMCID: PMC1287787
- DOI: 10.1128/JCM.43.11.5588-5592.2005
Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge
Abstract
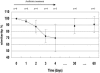
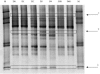
Recent studies have shown that the human fecal microbiota is composed of a consortium of species specific to the host and resistant to modifications over time. Antibiotics are known to affect the intestinal microflora, and ensuing changes may result in antibiotic-associated diarrhea. It is therefore important to characterize the nature and amplitude of these modifications and the ability of this ecosystem to return to its original profile-i.e., its resilience. Six healthy volunteers received oral amoxicillin (1.5 g/day) for 5 days. Fecal samples were collected at day 0 (D0) before antibiotic treatment and at set intervals until 60 days thereafter. Fecal DNA was isolated, and V6-to-V8 regions of the 16S rRNA genes were amplified by PCR with general primers and analyzed by temporal temperature gradient gel electrophoresis. Dominant species profiles were compared on the basis of similarity (Pearson correlation coefficient). Dominant species profiles at D0 were used as a reference. The fecal microbiota showed a major shift in dominant species upon antibiotic treatment, starting 24 h after treatment initiation and reaching an average similarity of only 74% after 4 days. Within 30 days following antibiotic treatment, the fecal microbiota tended to reach an average similarity of 88% to the D0 value; within 60 days, the average similarity to the D0 value was 89%. However, in one subject, important modifications persisted for at least 2 months, with similarity to the D0 value remaining below 70%. We demonstrated the resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. Yet the persistence of long-term alterations in some subjects may explain susceptibilities to antibiotic-associated diarrhea. Furthermore, these findings suggest that strategies reinforcing the ability of the fecal microbiota to resist modifications would be of clinical relevance.
Figures
References
-
- Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. - PMC - PubMed
-
- Beaugerie, L., and J. C. Petit. 2004. Microbial-gut interactions in health and disease. Antibiotic-associated diarrhoea. Best Pract. Res. Clin. Gastroenterol. 18:337-352. - PubMed
-
- Bonnet, R., A. Suau, J. Dore, G. R. Gibson, and M. D. Collins. 2002. Differences in rDNA libraries of faecal bacteria derived from 10- and 25-cycle PCRs. Int. J. Syst. Evol. Microbiol. 52:757-763. - PubMed
-
- Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. - PubMed
-
- Falk, P., L. C. Hoskins, and G. Larson. 1990. Bacteria of the human intestinal microbiota produce glycosidases specific for lacto-series glycosphingolipids. J. Biochem. (Tokyo) 108:466-474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

