Molecular epidemiology of clinical Cryptococcus neoformans strains from India
- PMID: 16272511
- PMCID: PMC1287776
- DOI: 10.1128/JCM.43.11.5733-5742.2005
Molecular epidemiology of clinical Cryptococcus neoformans strains from India
Abstract
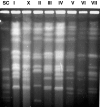
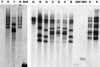
Little is known about the molecular epidemiology of the human pathogenic fungus Cryptococcus neoformans in India, a country now in the midst of an epidemic of AIDS-related cryptococcosis. We studied 57 clinical isolates from several regions in India, of which 51 were C. neoformans var. grubii, 1 was C. neoformans var. neoformans, and 5 were C. neoformans var. gattii. This strain set included 18 additional sequential isolates from 14 patients. Strains were characterized phenotypically by measuring the polysaccharide capsule and by determining the MICs of standard antifungals. Molecular typing was performed by a PCR-based method using the minisatellite-specific core sequence (M13), by electrophoretic karyotyping, by restriction fragment length polymorphisms with the C. neoformans transposon 1 (TCN-1), and by URA5 DNA sequence analysis. Overall, Indian isolates were less heterogeneous than isolates from other regions and included a subset that clustered into one group based on URA5 DNA sequence analysis. In summary, our results demonstrate (i) differences in genetic diversity of C. neoformans isolates from India compared to isolates from other regions in the world; (ii) that DNA typing with the TCN-1 probe can adequately distinguish C. neoformans var. grubii strains; (iii) that TCN-1 sequences are absent in many C. neoformans var. gattii strains, supporting previous studies indicating that these strains have a limited geographical dispersal; and (iv) that human cryptococcal infection can be associated with microevolution of the infecting strain and by simultaneous coinfection with two distinct C. neoformans strains.
Figures


Similar articles
-
In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from north-western India.J Med Microbiol. 2011 Jul;60(Pt 7):961-967. doi: 10.1099/jmm.0.029025-0. Epub 2011 Mar 10. J Med Microbiol. 2011. PMID: 21393452
-
Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand.PLoS Negl Trop Dis. 2013 Jul 4;7(7):e2297. doi: 10.1371/journal.pntd.0002297. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23861989 Free PMC article.
-
Molecular typing of clinical Cryptococcus neoformans isolates collected in Germany from 2004 to 2010.Med Microbiol Immunol. 2014 Oct;203(5):333-40. doi: 10.1007/s00430-014-0341-6. Epub 2014 May 17. Med Microbiol Immunol. 2014. PMID: 24838744
-
Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII.Mycopathologia. 2012 Jun;173(5-6):337-46. doi: 10.1007/s11046-011-9491-x. Epub 2011 Nov 13. Mycopathologia. 2012. PMID: 22081254
-
Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure.J Clin Microbiol. 1997 Sep;35(9):2243-51. doi: 10.1128/jcm.35.9.2243-2251.1997. J Clin Microbiol. 1997. PMID: 9276395 Free PMC article.
Cited by
-
Biofilm formation in clinical Candida isolates and its association with virulence.Microbes Infect. 2009 Jul-Aug;11(8-9):753-61. doi: 10.1016/j.micinf.2009.04.018. Epub 2009 May 4. Microbes Infect. 2009. PMID: 19409507 Free PMC article.
-
Distribution and association between environmental and clinical isolates of Cryptococcus neoformans in Bogotá-Colombia, 2012-2015.Mem Inst Oswaldo Cruz. 2016 Oct;111(10):642-648. doi: 10.1590/0074-02760160201. Epub 2016 Oct 3. Mem Inst Oswaldo Cruz. 2016. Retraction in: Mem Inst Oswaldo Cruz. 2017 Nov;112(11):796. doi: 10.1590/0074-02760160560. PMID: 27706379 Free PMC article. Retracted.
-
Ecoepidemiology of Cryptococcus gattii in Developing Countries.J Fungi (Basel). 2017 Nov 3;3(4):62. doi: 10.3390/jof3040062. J Fungi (Basel). 2017. PMID: 29371578 Free PMC article. Review.
-
Characterization of the virulence of Cryptococcus neoformans strains in an insect model.Virulence. 2015;6(8):809-13. doi: 10.1080/21505594.2015.1086868. Epub 2015 Sep 12. Virulence. 2015. PMID: 26364757 Free PMC article. No abstract available.
-
Abdominal Lymphonodular Cryptococcosis in an Immunocompetent Child.Case Rep Pediatr. 2015;2015:347403. doi: 10.1155/2015/347403. Epub 2015 Nov 16. Case Rep Pediatr. 2015. PMID: 26649217 Free PMC article.
References
-
- Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. - PubMed
-
- Banerjee, U., K. Datta, and A. Casadevall. 2004. Serotype distribution of Cryptococcus neoformans in patients in a tertiary care center in India. Med. Mycol. 42:181-186. - PubMed
-
- Banerjee, U., K. Datta, T. Majumdar, and K. Gupta. 2001. Cryptococcosis in India: the awakening of a giant? Med. Mycol. 39:51-67. - PubMed
-
- Banerjee, U., K. Gupta, B. Chatterjee, and S. Sethi. 1994. Cryptococcosis at AIIMS. Natl. Med. J. India 7:51-52. - PubMed
-
- Banerjee, U., J. B. Khadka, S. Sethi, and K. Gupta. 1995. Sudden spurt of cryptococcosis at a tertiary care hospital at New Delhi between December 1994 to February 1995. Indian J. Med. Res. 102:272-274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

