Structural basis for transcription inhibition by tagetitoxin
- PMID: 16273103
- PMCID: PMC1790907
- DOI: 10.1038/nsmb1015
Structural basis for transcription inhibition by tagetitoxin
Abstract
Tagetitoxin (Tgt) inhibits transcription by an unknown mechanism. A structure at a resolution of 2.4 A of the Thermus thermophilus RNA polymerase (RNAP)-Tgt complex revealed that the Tgt-binding site within the RNAP secondary channel overlaps that of the stringent control effector ppGpp, which partially protects RNAP from Tgt inhibition. Tgt binding is mediated exclusively through polar interactions with the beta and beta' residues whose substitutions confer resistance to Tgt in vitro. Importantly, a Tgt phosphate, together with two active site acidic residues, coordinates the third Mg(2+) ion, which is distinct from the two catalytic metal ions. We show that Tgt inhibits all RNAP catalytic reactions and propose a mechanism in which the Tgt-bound Mg(2+) ion has a key role in stabilization of an inactive transcription intermediate. Remodeling of the active site by metal ions could be a common theme in the regulation of catalysis by nucleic acid enzymes.
Figures

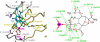
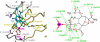
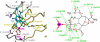



Similar articles
-
Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop.J Biol Chem. 2011 Nov 18;286(46):40395-400. doi: 10.1074/jbc.M111.300889. Epub 2011 Oct 5. J Biol Chem. 2011. PMID: 21976682 Free PMC article.
-
Response to Klyuyev and Vassylyev: on the mechanism of tagetitoxin inhibition of transcription.Transcription. 2012 Mar-Apr;3(2):51-5. doi: 10.4161/trns.19749. Epub 2012 Mar 1. Transcription. 2012. PMID: 22414748 Free PMC article.
-
Active site opening and closure control translocation of multisubunit RNA polymerase.Nucleic Acids Res. 2012 Aug;40(15):7442-51. doi: 10.1093/nar/gks383. Epub 2012 May 8. Nucleic Acids Res. 2012. PMID: 22570421 Free PMC article.
-
Bacterial RNA polymerase inhibitors: an organized overview of their structure, derivatives, biological activity and current clinical development status.Curr Med Chem. 2009;16(4):430-54. doi: 10.2174/092986709787315559. Curr Med Chem. 2009. PMID: 19199915 Review.
-
[Low-molecular weight inhibitors of bacterial DNA-dependent RNA polymerase].Mol Biol (Mosk). 2006 Nov-Dec;40(6):971-81. Mol Biol (Mosk). 2006. PMID: 17209424 Review. Russian.
Cited by
-
Bacterial Transcription as a Target for Antibacterial Drug Development.Microbiol Mol Biol Rev. 2016 Jan 13;80(1):139-60. doi: 10.1128/MMBR.00055-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26764017 Free PMC article. Review.
-
Ideality in Context: Motivations for Total Synthesis.Acc Chem Res. 2021 Feb 2;54(3):605-617. doi: 10.1021/acs.accounts.0c00821. Epub 2021 Jan 21. Acc Chem Res. 2021. PMID: 33476518 Free PMC article.
-
Active site closure stabilizes the backtracked state of RNA polymerase.Nucleic Acids Res. 2018 Nov 16;46(20):10870-10887. doi: 10.1093/nar/gky883. Nucleic Acids Res. 2018. PMID: 30256972 Free PMC article.
-
Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis.Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):10878-83. doi: 10.1073/pnas.0914424107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534498 Free PMC article.
-
An allosteric mechanism of Rho-dependent transcription termination.Nature. 2010 Jan 14;463(7278):245-9. doi: 10.1038/nature08669. Nature. 2010. PMID: 20075920 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

