Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts
- PMID: 16273343
- PMCID: PMC1544375
- DOI: 10.1007/s00122-005-0112-2
Coffee and tomato share common gene repertoires as revealed by deep sequencing of seed and cherry transcripts
Abstract
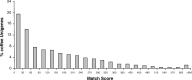
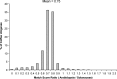
An EST database has been generated for coffee based on sequences from approximately 47,000 cDNA clones derived from five different stages/tissues, with a special focus on developing seeds. When computationally assembled, these sequences correspond to 13,175 unigenes, which were analyzed with respect to functional annotation, expression profile and evolution. Compared with Arabidopsis, the coffee unigenes encode a higher proportion of proteins related to protein modification/turnover and metabolism-an observation that may explain the high diversity of metabolites found in coffee and related species. Several gene families were found to be either expanded or unique to coffee when compared with Arabidopsis. A high proportion of these families encode proteins assigned to functions related to disease resistance. Such families may have expanded and evolved rapidly under the intense pathogen pressure experienced by a tropical, perennial species like coffee. Finally, the coffee gene repertoire was compared with that of Arabidopsis and Solanaceous species (e.g. tomato). Unlike Arabidopsis, tomato has a nearly perfect gene-for-gene match with coffee. These results are consistent with the facts that coffee and tomato have a similar genome size, chromosome karyotype (tomato, n=12; coffee n=11) and chromosome architecture. Moreover, both belong to the Asterid I clade of dicot plant families. Thus, the biology of coffee (family Rubiacaeae) and tomato (family Solanaceae) may be united into one common network of shared discoveries, resources and information.
Figures





References
-
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. - PubMed
-
- Chase MW, Soltis DE, et al. Phylogenetics of seed plants—an analysis of nucleotide-sequences from the plastid gene rbcl. Ann Mo Bot Gard. 1993;80:528–580. doi: 10.2307/2399846. - DOI
-
- Chen ZJ, Ribeiro R, Silva MC. Heat shock-induced susceptibility of green coffee leaves and berries to Colletotrichum gloeosporioides and its association to PR and hsp70 gene expression. Physiol Mol Plant P. 2003;63:181–190. doi: 10.1016/j.pmpp.2003.11.004. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

