Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon
- PMID: 16278345
- PMCID: PMC1315375
- DOI: 10.1105/tpc.105.036889
Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon
Abstract
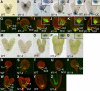
Pattern formation and morphogenesis require coordination of cell division rates and orientations with developmental signals that specify cell fate. A viable mutation in the TILTED1 locus, which encodes the catalytic subunit of DNA polymerase epsilon of Arabidopsis thaliana, causes a lengthening of the cell cycle by approximately 35% throughout embryo development and alters cell type patterning of the hypophyseal lineage in the root, leading to a displacement of the root pole from its normal position on top of the suspensor. Treatment of preglobular and early globular stages, but not later stage, embryos with the DNA polymerase inhibitor aphidicolin leads to a similar phenotype. The results uncover an interaction between the cell cycle and the processes that determine cell fate during plant embryogenesis.
Figures




References
-
- Aida, M., Beis, D., Heidstra, H., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.-S., Amasino, R.M., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. - PubMed
-
- Alonso, J.M.A., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Ambros, V. (1999). Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development 126 1947–1956. - PubMed
-
- Berleth, T., and Jürgens, G. (1993). The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118 575–587.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

