Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica
- PMID: 16278444
- PMCID: PMC1287852
- DOI: 10.1128/EC.4.11.1775-1784.2005
Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica
Abstract
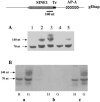
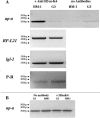
Transcriptional silencing of an amoebapore (ap-a) gene occurred in Entamoeba histolytica following the transfection of plasmids containing a DNA segment (473 bp) homologous to the 5' upstream region of the gene (R. Bracha, Y. Nuchamowitz, and D. Mirelman, Eukaryot. Cell 2:295-305, 2003). This segment contains the promoter region of the ap-a gene, a T-rich stretch, followed by a truncated SINE1 (short interspersed element 1) that is transcribed from the antisense strand. Transfection of plasmids containing truncated SINE1 sequences which lack their 3' regulatory elements upstream of the ap-a gene was essential for the downstream silencing of the ap-a gene while transfection with plasmids containing the entire SINE1 sequence or without the T-rich stretch promoted the overexpression of the ap-a gene. Both the T-rich stretch and sequences of the 5' SINE1 were essential for the transcription of SINE1. RNA extracts from gene-silenced cultures showed small amounts of short (approximately 140-nucleotide), single-stranded molecules with homology to SINE1 but no short interfering RNA. Chromatin immunoprecipitation analysis with an antibody against methylated K4 of histone H3 showed a demethylation of K4 at the domain of the ap-a gene, indicating transcriptional inactivation. These results suggest the involvement of SINE1 in triggering the gene silencing and the role of histone modification in its epigenetic maintenance.
Figures




References
-
- Abed, M., and S. Ankri. 2005. Molecular characterization of Entamoeba histolytica RNase III and AGO2, two RNA interference hallmark proteins. Exp. Parasitol. 110:265-269. - PubMed
-
- Alon, R. N., R. Bracha, and D. Mirelman. 1997. Inhibition of expression of the lysine-rich 30 kDa surface antigen of Entamoeba dispar by the transcription of its antisense RNA. Mol. Biochem. Parasitol. 90:193-201. - PubMed
-
- Bakrea, A. A., K. Rawal, R. Ramaswamy, A. Bhattacharya, and S. Bhattacharya. 2005. The LINEs and SINEs of Entamoeba histolytica: comparative analysis and genomic distribution. Exp. Parasitol. 110:207-213. - PubMed
-
- Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

