Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae
- PMID: 16278453
- PMCID: PMC1287855
- DOI: 10.1128/EC.4.11.1863-1871.2005
Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae
Abstract
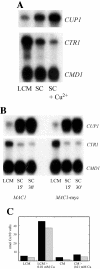
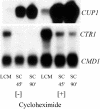
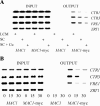
Ace1 and Mac1 undergo reciprocal copper metalloregulation in yeast cells. Mac1 is functional as a transcriptional activator in copper-deficient cells, whereas Ace1 is a transcriptional activator in copper-replete cells. Cells undergoing a transition from copper-deficient to copper-sufficient conditions through a switch in the growth medium show a rapid inactivation of Mac1 and a corresponding rise in Ace1 activation. Cells analyzed after the transition show a massive accumulation of cellular copper. Under these copper shock conditions we show, using two epitope-tagged variants of Mac1, that copper-mediated inhibition of Mac function is independent of induced protein turnover. The transcription activity of Mac1 is rapidly inhibited in the copper-replete cells, whereas chromatin immunoprecipitation studies showed only partial copper-induced loss of DNA binding. Thus, the initial event in copper inhibition of Mac1 function is likely copper inhibition of the transactivation activity. Copper inhibition of Mac1 in transition experiments is largely unaffected in cells overexpressing copper-binding proteins within the nucleus. Likewise, high expression of a copper-binding, non-DNA-binding Mac1 mutant is without effect on the copper activation of Ace1. Thus, metalloregulation of Ace1 and Mac1 occurs independently.
Figures





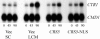
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current Protocols in Molecular Biology, vol. 2. John Wiley & Sons, New York, N.Y.
-
- Bloomfield, S. F., G. S. A. B. Stewart, C. E. R. Dodd, I. R. Booth, and E. G. M. Power. 1998. The viable but non-culturable phenomenon explained? Microbiology 144:1-3. - PubMed
-
- Brown, K. R., G. L. Keller, I. J. Pickering, H. H. Harris, G. N. George, and D. R. Winge. 2002. Structures of the cuprous-thiolate clusters of the Mac1 and Ace1 transcriptional activators. Biochemistry 41:6469-6476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

