A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans
- PMID: 16278457
- PMCID: PMC1287864
- DOI: 10.1128/EC.4.11.1902-1912.2005
A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans
Abstract
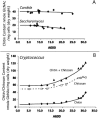
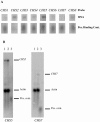
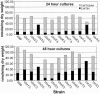
Chitin is an essential component of the cell wall of many fungi. Chitin also can be enzymatically deacetylated to chitosan, a more flexible and soluble polymer. Cryptococcus neoformans is a fungal pathogen that causes cryptococcal meningoencephalitis, particularly in immunocompromised patients. In this work, we show that both chitin and chitosan are present in the cell wall of vegetatively growing C. neoformans yeast cells and that the levels of both rise dramatically as cells grow to higher density in liquid culture. C. neoformans has eight putative chitin synthases, and strains with any one chitin synthase deleted are viable at 30 degrees C. In addition, C. neoformans genes encode three putative regulator proteins, which are homologs of Saccharomyces cerevisiae Skt5p. None of these three is essential for viability. However, one of the chitin synthases (Chs3) and one of the regulators (Csr2) are important for growth. Cells with deletions in either CHS3 or CSR2 have several shared phenotypes, including sensitivity to growth at 37 degrees C. The similarity of their phenotypes also suggests that Csr2 specifically regulates chitin synthesis by Chs3. Lastly, both chs3Delta and the csr2Delta mutants are defective in chitosan production, predicting that Chs3-Csr2 complex with chitin deacetylases for conversion of chitin to chitosan. These data suggest that chitin synthesis could be an excellent antifungal target.
Figures


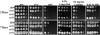


References
-
- Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5:217-238. - PubMed
-
- Araki, Y., and E. Ito. 1975. A pathway of chitosan formation in Mucor rouxii. Enzymatic deacetylation of chitin. Eur. J. Biochem. 55:71-78. - PubMed
-
- Bartnicki-Garcia, S., and E. Lippman.1969. Fungal morphogenesis: cell wall construction in Mucor rouxii. Science 165:302-304. - PubMed
-
- Bartnicki-Garcia, S., and W. J. Nickerson. 1962. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim. Biophys. Acta 58:102-119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

